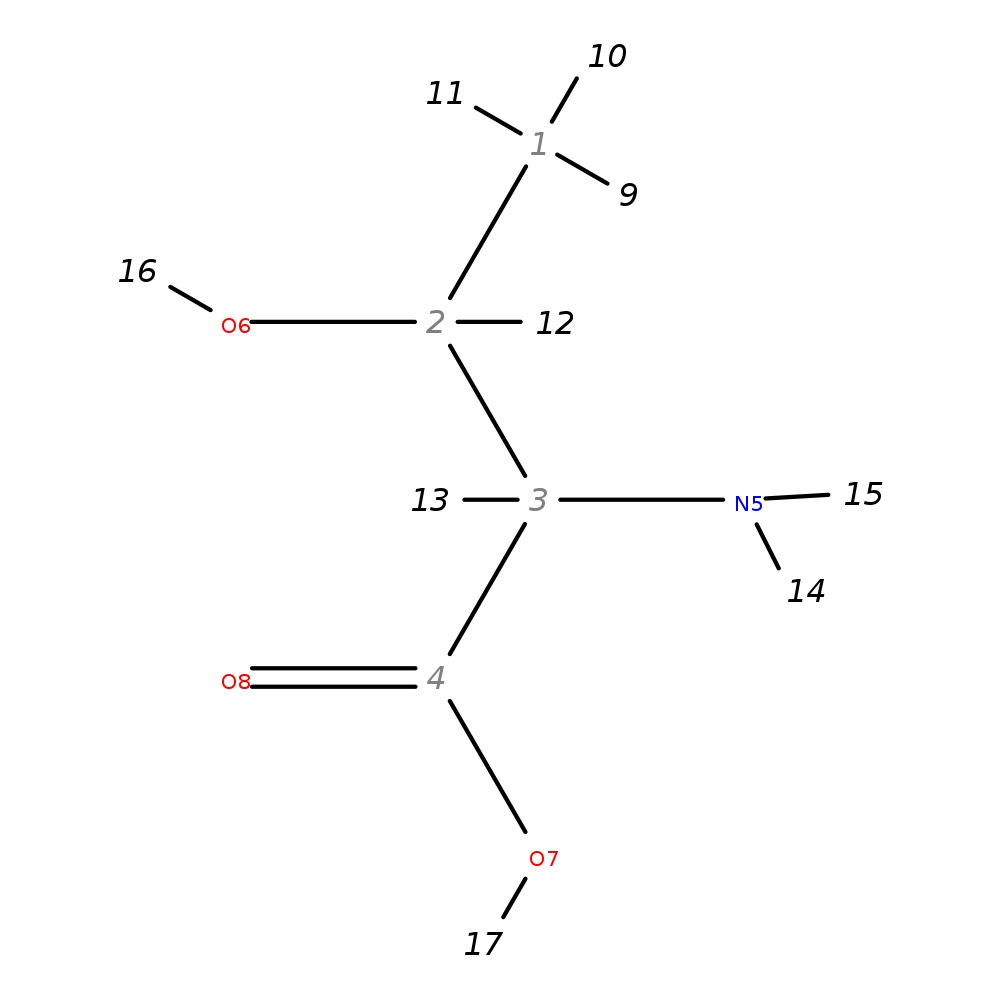
2-Amino-3-hydroxybutanoic acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01274 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C4H9NO3/c1-2(6)3(5)4(7)8/h2-3,6H,5H2,1H3,(H,7,8)/t2-,3-/m0/s1 | |
| Note 1 | Oscar Li |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-amino-3-hydroxybutanoic acid | solute | 100mM |
| D2O | solvent | 100.0% |
| DSS | reference | 0.01mg/mL |
| sodium phosphate | buffer | 50mM |
| sodium azide | cytocide | 500uM |
Spin System Matrix
| 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|
| 9 | 1.197 | -12.4 | 0 | 6.619 | 0 |
| 10 | 0 | 1.197 | 0 | 6.619 | 0 |
| 11 | 0 | 0 | 1.197 | 6.619 | 0 |
| 12 | 0 | 0 | 0 | 4.354 | 3.92 |
| 13 | 0 | 0 | 0 | 0 | 3.828 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.19023 | 1.0 | standard |
| 1.20349 | 0.998626 | standard |
| 3.82393 | 0.3379 | standard |
| 3.83176 | 0.3379 | standard |
| 4.33008 | 0.0439795 | standard |
| 4.33794 | 0.0493339 | standard |
| 4.34336 | 0.130242 | standard |
| 4.35121 | 0.133793 | standard |
| 4.35658 | 0.13359 | standard |
| 4.36442 | 0.13006 | standard |
| 4.36981 | 0.049406 | standard |
| 4.37767 | 0.0439855 | standard |
| 1.11297 | 0.893579 | standard |
| 1.27649 | 1.0 | standard |
| 3.77434 | 0.268958 | standard |
| 3.87182 | 0.391998 | standard |
| 4.07171 | 0.0668021 | standard |
| 4.17017 | 0.044631 | standard |
| 4.23185 | 0.138864 | standard |
| 4.33005 | 0.0984831 | standard |
| 4.3958 | 0.124954 | standard |
| 4.494 | 0.0909062 | standard |
| 4.56199 | 0.0463291 | standard |
| 4.66005 | 0.03266 | standard |
| 1.14102 | 0.932309 | standard |
| 1.25081 | 1.0 | standard |
| 3.79317 | 0.292413 | standard |
| 3.85836 | 0.373302 | standard |
| 4.16159 | 0.0559349 | standard |
| 4.22726 | 0.047195 | standard |
| 4.27076 | 0.137966 | standard |
| 4.33621 | 0.111562 | standard |
| 4.38052 | 0.131221 | standard |
| 4.44593 | 0.104206 | standard |
| 4.4907 | 0.047534 | standard |
| 4.55625 | 0.0356384 | standard |
| 1.15508 | 0.953976 | standard |
| 1.23759 | 1.00004 | standard |
| 3.80221 | 0.303772 | standard |
| 3.85115 | 0.364063 | standard |
| 4.2084 | 0.0521601 | standard |
| 4.2576 | 0.0480399 | standard |
| 4.29072 | 0.13637 | standard |
| 4.33976 | 0.117293 | standard |
| 4.37321 | 0.134872 | standard |
| 4.42225 | 0.112083 | standard |
| 4.45575 | 0.0481861 | standard |
| 4.50489 | 0.0374176 | standard |
| 1.15976 | 0.961678 | standard |
| 1.23313 | 1.00004 | standard |
| 3.80515 | 0.307732 | standard |
| 3.84864 | 0.361314 | standard |
| 4.22417 | 0.0510792 | standard |
| 4.26788 | 0.048278 | standard |
| 4.29746 | 0.135909 | standard |
| 4.34105 | 0.119097 | standard |
| 4.37077 | 0.135816 | standard |
| 4.41443 | 0.11507 | standard |
| 4.44417 | 0.0484359 | standard |
| 4.48783 | 0.0380777 | standard |
| 1.1635 | 0.965846 | standard |
| 1.22956 | 1.00005 | standard |
| 3.80753 | 0.310565 | standard |
| 3.84667 | 0.358707 | standard |
| 4.23697 | 0.0502083 | standard |
| 4.27629 | 0.0484624 | standard |
| 4.30289 | 0.135296 | standard |
| 4.34219 | 0.120842 | standard |
| 4.36896 | 0.136002 | standard |
| 4.40817 | 0.116499 | standard |
| 4.43496 | 0.0485341 | standard |
| 4.47435 | 0.0385655 | standard |
| 1.18024 | 1.00008 | standard |
| 1.21334 | 0.999139 | standard |
| 3.81788 | 0.327349 | standard |
| 3.8374 | 0.349545 | standard |
| 4.29488 | 0.0457713 | standard |
| 4.31448 | 0.048005 | standard |
| 4.32796 | 0.133764 | standard |
| 4.34751 | 0.129214 | standard |
| 4.36096 | 0.136901 | standard |
| 4.38062 | 0.125839 | standard |
| 4.39397 | 0.0508207 | standard |
| 4.41367 | 0.0426645 | standard |
| 1.18579 | 1.00018 | standard |
| 1.20788 | 0.999361 | standard |
| 3.82125 | 0.335514 | standard |
| 3.83423 | 0.340923 | standard |
| 4.31441 | 0.0451034 | standard |
| 4.32755 | 0.0484585 | standard |
| 4.33649 | 0.130745 | standard |
| 4.34952 | 0.13255 | standard |
| 4.35849 | 0.133554 | standard |
| 4.3716 | 0.13011 | standard |
| 4.3805 | 0.050482 | standard |
| 4.39364 | 0.0430785 | standard |
| 1.18857 | 1.00026 | standard |
| 1.20513 | 1.00026 | standard |
| 3.82285 | 0.338093 | standard |
| 3.83265 | 0.338264 | standard |
| 4.32423 | 0.0440395 | standard |
| 4.33408 | 0.0495724 | standard |
| 4.34075 | 0.129932 | standard |
| 4.35055 | 0.133804 | standard |
| 4.35727 | 0.13379 | standard |
| 4.3671 | 0.130299 | standard |
| 4.37377 | 0.0495647 | standard |
| 4.38362 | 0.0440375 | standard |
| 1.19023 | 1.0 | standard |
| 1.20349 | 0.998626 | standard |
| 3.82393 | 0.337897 | standard |
| 3.83176 | 0.337897 | standard |
| 4.33008 | 0.0439785 | standard |
| 4.33794 | 0.0493298 | standard |
| 4.34336 | 0.130239 | standard |
| 4.35121 | 0.133788 | standard |
| 4.35658 | 0.133584 | standard |
| 4.36442 | 0.130057 | standard |
| 4.36981 | 0.049401 | standard |
| 4.37767 | 0.0439845 | standard |
| 1.19133 | 1.00017 | standard |
| 1.2024 | 0.999335 | standard |
| 3.82452 | 0.338313 | standard |
| 3.83106 | 0.338313 | standard |
| 4.33404 | 0.0440394 | standard |
| 4.34061 | 0.049517 | standard |
| 4.34508 | 0.130145 | standard |
| 4.35163 | 0.133727 | standard |
| 4.35608 | 0.133727 | standard |
| 4.36264 | 0.130119 | standard |
| 4.36714 | 0.0494317 | standard |
| 4.37371 | 0.0440304 | standard |
| 1.19211 | 1.0 | standard |
| 1.20161 | 0.997299 | standard |
| 3.82502 | 0.338011 | standard |
| 3.83057 | 0.338011 | standard |
| 4.33692 | 0.0440105 | standard |
| 4.34249 | 0.0494272 | standard |
| 4.34634 | 0.130276 | standard |
| 4.35193 | 0.13334 | standard |
| 4.35579 | 0.133316 | standard |
| 4.36141 | 0.130239 | standard |
| 4.36526 | 0.0494191 | standard |
| 4.37083 | 0.0440085 | standard |
| 1.19244 | 1.0009 | standard |
| 1.20126 | 1.0009 | standard |
| 3.82521 | 0.339154 | standard |
| 3.83047 | 0.339154 | standard |
| 4.33801 | 0.0441525 | standard |
| 4.34328 | 0.0497368 | standard |
| 4.34683 | 0.130855 | standard |
| 4.35203 | 0.133763 | standard |
| 4.35566 | 0.134228 | standard |
| 4.36085 | 0.130322 | standard |
| 4.36447 | 0.0495234 | standard |
| 4.36974 | 0.0441324 | standard |
| 1.19271 | 1.0 | standard |
| 1.20104 | 1.0 | standard |
| 3.82541 | 0.338023 | standard |
| 3.83027 | 0.338023 | standard |
| 4.339 | 0.0440035 | standard |
| 4.34388 | 0.0493522 | standard |
| 4.34726 | 0.129688 | standard |
| 4.35219 | 0.133758 | standard |
| 4.35556 | 0.133817 | standard |
| 4.36042 | 0.130329 | standard |
| 4.36377 | 0.0494495 | standard |
| 4.36875 | 0.0439524 | standard |
| 1.1932 | 1.0 | standard |
| 1.20054 | 1.0 | standard |
| 3.82561 | 0.337837 | standard |
| 3.82997 | 0.337837 | standard |
| 4.34059 | 0.0439524 | standard |
| 4.34497 | 0.04917 | standard |
| 4.34802 | 0.130245 | standard |
| 4.35233 | 0.132993 | standard |
| 4.35536 | 0.133607 | standard |
| 4.35973 | 0.130266 | standard |
| 4.36268 | 0.0495257 | standard |
| 4.36706 | 0.0439815 | standard |
| 1.1934 | 1.0 | standard |
| 1.20034 | 1.0 | standard |
| 3.82571 | 0.337726 | standard |
| 3.82987 | 0.337726 | standard |
| 4.34128 | 0.0439414 | standard |
| 4.34546 | 0.0493044 | standard |
| 4.34832 | 0.130296 | standard |
| 4.35239 | 0.133701 | standard |
| 4.35526 | 0.133669 | standard |
| 4.35936 | 0.129539 | standard |
| 4.36219 | 0.0494305 | standard |
| 4.36637 | 0.0439504 | standard |
| 1.1936 | 1.0 | standard |
| 1.20015 | 1.0 | standard |
| 3.82591 | 0.338003 | standard |
| 3.82977 | 0.338003 | standard |
| 4.34198 | 0.0440146 | standard |
| 4.34586 | 0.0494084 | standard |
| 4.34855 | 0.129382 | standard |
| 4.35249 | 0.133794 | standard |
| 4.35516 | 0.13376 | standard |
| 4.35913 | 0.130205 | standard |
| 4.36179 | 0.0495355 | standard |
| 4.36568 | 0.0440236 | standard |
| 1.1939 | 1.0 | standard |
| 1.19985 | 1.0 | standard |
| 3.82601 | 0.33779 | standard |
| 3.82958 | 0.33779 | standard |
| 4.34307 | 0.0439995 | standard |
| 4.34665 | 0.0497909 | standard |
| 4.34902 | 0.130333 | standard |
| 4.35259 | 0.133614 | standard |
| 4.35506 | 0.133606 | standard |
| 4.35864 | 0.130181 | standard |
| 4.3611 | 0.0493047 | standard |
| 4.36468 | 0.0439555 | standard |
| 1.1943 | 1.0 | standard |
| 1.19945 | 1.0 | standard |
| 3.8263 | 0.337674 | standard |
| 3.82938 | 0.337674 | standard |
| 4.34465 | 0.0439304 | standard |
| 4.34774 | 0.0494081 | standard |
| 4.34981 | 0.13036 | standard |
| 4.35279 | 0.133837 | standard |
| 4.35486 | 0.133837 | standard |
| 4.35784 | 0.13036 | standard |
| 4.35991 | 0.0494081 | standard |
| 4.363 | 0.0439304 | standard |