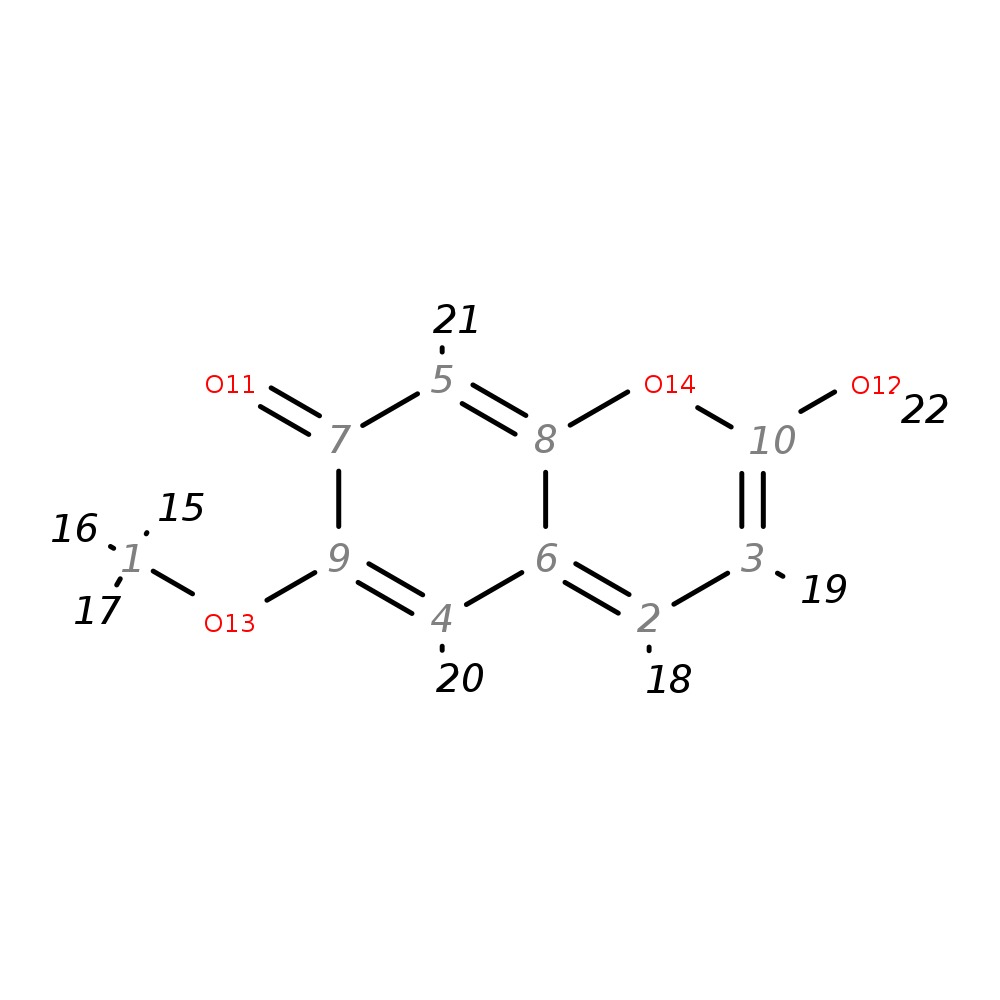
Scopoletin
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01302 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H8O4/c1-13-9-4-6-2-3-10(12)14-8(6)5-7(9)11/h2-5,12H,1H3 | |
| Note 1 | Oscar Li |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Scopoletin | solute | 100mM |
| DMSO | solvent | 100.0% |
| TMS | reference | 0.05% |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|
| 15 | 3.824 | -12.4 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 3.824 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 3.824 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 7.919 | 9.406 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 6.229 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 6.789 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 7.227 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.82372 | 1.0 | standard |
| 6.21908 | 0.167459 | standard |
| 6.23792 | 0.167459 | standard |
| 6.7894 | 0.333338 | standard |
| 7.22671 | 0.333338 | standard |
| 7.90971 | 0.167458 | standard |
| 7.92855 | 0.167458 | standard |
| 3.82372 | 1.0 | standard |
| 6.10284 | 0.144909 | standard |
| 6.338 | 0.1909 | standard |
| 6.7894 | 0.334188 | standard |
| 7.22671 | 0.33408 | standard |
| 7.80963 | 0.190676 | standard |
| 8.04479 | 0.144805 | standard |
| 3.82372 | 1.0 | standard |
| 6.14657 | 0.152292 | standard |
| 6.30328 | 0.183007 | standard |
| 6.7894 | 0.333701 | standard |
| 7.22671 | 0.333659 | standard |
| 7.84435 | 0.182921 | standard |
| 8.00105 | 0.152241 | standard |
| 3.82372 | 1.0 | standard |
| 6.1677 | 0.156037 | standard |
| 6.28533 | 0.179089 | standard |
| 6.7894 | 0.333538 | standard |
| 7.22671 | 0.333515 | standard |
| 7.8623 | 0.179045 | standard |
| 7.97993 | 0.156007 | standard |
| 3.82372 | 1.0 | standard |
| 6.17474 | 0.157296 | standard |
| 6.27918 | 0.17779 | standard |
| 6.7894 | 0.333494 | standard |
| 7.22671 | 0.333477 | standard |
| 7.86845 | 0.177754 | standard |
| 7.97298 | 0.15727 | standard |
| 3.82372 | 1.0 | standard |
| 6.1802 | 0.158301 | standard |
| 6.27432 | 0.176748 | standard |
| 6.7894 | 0.333464 | standard |
| 7.22671 | 0.33345 | standard |
| 7.87341 | 0.176723 | standard |
| 7.96743 | 0.158283 | standard |
| 3.82372 | 1.0 | standard |
| 6.20469 | 0.165179 | standard |
| 6.25171 | 0.169795 | standard |
| 6.7894 | 0.333387 | standard |
| 7.22671 | 0.333384 | standard |
| 7.89592 | 0.169788 | standard |
| 7.94293 | 0.165174 | standard |
| 3.82372 | 1.0 | standard |
| 6.21273 | 0.167466 | standard |
| 6.24407 | 0.167467 | standard |
| 6.7894 | 0.333348 | standard |
| 7.22671 | 0.333346 | standard |
| 7.90356 | 0.167464 | standard |
| 7.9349 | 0.167464 | standard |
| 3.82372 | 1.0 | standard |
| 6.21669 | 0.167463 | standard |
| 6.2402 | 0.167464 | standard |
| 6.7894 | 0.333341 | standard |
| 7.22671 | 0.333341 | standard |
| 7.90743 | 0.167462 | standard |
| 7.93093 | 0.167462 | standard |
| 3.82372 | 1.0 | standard |
| 6.21908 | 0.167459 | standard |
| 6.23792 | 0.167459 | standard |
| 6.7894 | 0.333338 | standard |
| 7.22671 | 0.333338 | standard |
| 7.90971 | 0.167458 | standard |
| 7.92855 | 0.167458 | standard |
| 3.82372 | 1.0 | standard |
| 6.22066 | 0.167461 | standard |
| 6.23633 | 0.167461 | standard |
| 6.7894 | 0.333337 | standard |
| 7.22671 | 0.333337 | standard |
| 7.91129 | 0.167461 | standard |
| 7.92696 | 0.167461 | standard |
| 3.82372 | 1.0 | standard |
| 6.22185 | 0.167466 | standard |
| 6.23524 | 0.167466 | standard |
| 6.7894 | 0.333336 | standard |
| 7.22671 | 0.333336 | standard |
| 7.91238 | 0.167454 | standard |
| 7.92587 | 0.167454 | standard |
| 3.82372 | 1.0 | standard |
| 6.22225 | 0.167453 | standard |
| 6.23485 | 0.167453 | standard |
| 6.7894 | 0.333336 | standard |
| 7.22671 | 0.333335 | standard |
| 7.91288 | 0.167465 | standard |
| 7.92538 | 0.167465 | standard |
| 3.82372 | 1.0 | standard |
| 6.22265 | 0.167454 | standard |
| 6.23445 | 0.167454 | standard |
| 6.7894 | 0.333335 | standard |
| 7.22671 | 0.333335 | standard |
| 7.91318 | 0.167453 | standard |
| 7.92498 | 0.167453 | standard |
| 3.82372 | 1.0 | standard |
| 6.22334 | 0.167465 | standard |
| 6.23375 | 0.167465 | standard |
| 6.7894 | 0.333335 | standard |
| 7.22671 | 0.333335 | standard |
| 7.91387 | 0.167465 | standard |
| 7.92429 | 0.167465 | standard |
| 3.82372 | 1.0 | standard |
| 6.22364 | 0.167473 | standard |
| 6.23346 | 0.167473 | standard |
| 6.7894 | 0.333335 | standard |
| 7.22671 | 0.333335 | standard |
| 7.91417 | 0.167457 | standard |
| 7.92409 | 0.167456 | standard |
| 3.82372 | 1.0 | standard |
| 6.22384 | 0.167457 | standard |
| 6.23326 | 0.167457 | standard |
| 6.7894 | 0.333335 | standard |
| 7.22671 | 0.333334 | standard |
| 7.91437 | 0.167456 | standard |
| 7.92379 | 0.167456 | standard |
| 3.82372 | 1.0 | standard |
| 6.22423 | 0.167445 | standard |
| 6.23286 | 0.167445 | standard |
| 6.7894 | 0.333334 | standard |
| 7.22671 | 0.333334 | standard |
| 7.91486 | 0.167463 | standard |
| 7.92339 | 0.167463 | standard |
| 3.82372 | 1.0 | standard |
| 6.22493 | 0.167458 | standard |
| 6.23217 | 0.167458 | standard |
| 6.7894 | 0.333334 | standard |
| 7.22671 | 0.333334 | standard |
| 7.91546 | 0.167458 | standard |
| 7.9227 | 0.167458 | standard |