L-Asparagine
Simulation outputs:
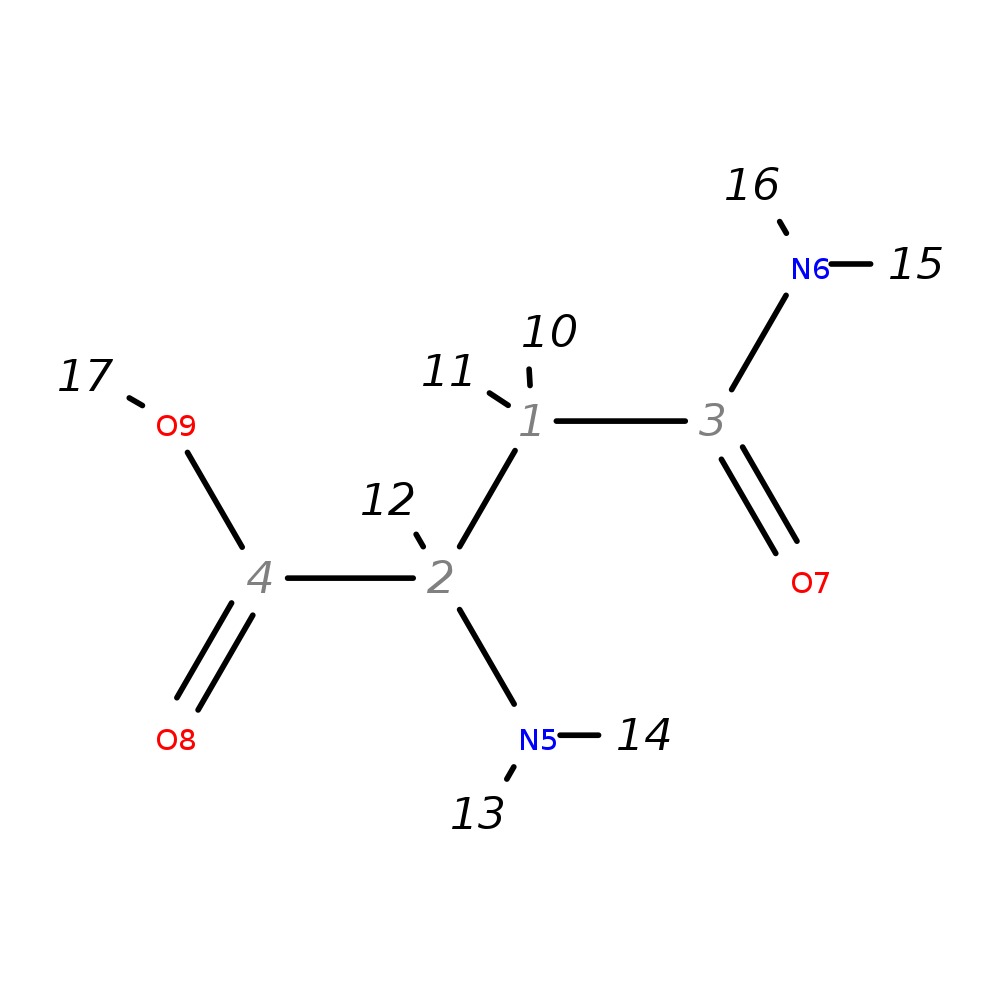
|
Parameter | Value |
| Field strength | 600.13(MHz) | |
| RMSD of the fit | 0.01952 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C4H8N2O3/c5-2(4(8)9)1-3(6)7/h2H,1,5H2,(H2,6,7)(H,8,9)/t2-/m0/s1 | |
| Note 1 | Copied spin matrix from bmse000030 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| L-Asparagine | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 10 | 11 | 12 | |
|---|---|---|---|
| 10 | 2.851 | -16.953 | 7.714 |
| 11 | 0 | 2.936 | 4.136 |
| 12 | 0 | 0 | 3.989 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.82829 | 0.515704 | standard |
| 2.84099 | 0.506873 | standard |
| 2.85656 | 0.968823 | standard |
| 2.86925 | 0.980464 | standard |
| 2.92094 | 0.985601 | standard |
| 2.92797 | 1.00002 | standard |
| 2.9492 | 0.527719 | standard |
| 2.95623 | 0.512101 | standard |
| 3.97906 | 0.761976 | standard |
| 3.9861 | 0.790904 | standard |
| 3.99174 | 0.790887 | standard |
| 3.99878 | 0.761976 | standard |
| 2.37684 | 0.00969613 | standard |
| 2.80103 | 0.477704 | standard |
| 2.82884 | 0.494478 | standard |
| 2.96194 | 1.00001 | standard |
| 3.25246 | 0.0189171 | standard |
| 3.5641 | 0.00865013 | standard |
| 3.85244 | 0.29342 | standard |
| 3.98271 | 0.272153 | standard |
| 4.01622 | 0.257831 | standard |
| 4.14678 | 0.176714 | standard |
| 2.54619 | 0.016769 | standard |
| 2.65794 | 0.0116432 | standard |
| 2.82888 | 0.477852 | standard |
| 2.85544 | 0.493566 | standard |
| 2.94049 | 1.0 | standard |
| 3.13785 | 0.022686 | standard |
| 3.89518 | 0.282741 | standard |
| 3.9818 | 0.281075 | standard |
| 4.00534 | 0.271846 | standard |
| 4.09206 | 0.200932 | standard |
| 2.62936 | 0.0292074 | standard |
| 2.71403 | 0.020566 | standard |
| 2.84125 | 0.540737 | standard |
| 2.86959 | 0.561744 | standard |
| 2.92926 | 1.0 | standard |
| 3.08134 | 0.0361462 | standard |
| 3.14465 | 0.0149104 | standard |
| 3.91769 | 0.324796 | standard |
| 3.98153 | 0.328834 | standard |
| 4.00161 | 0.319676 | standard |
| 4.0655 | 0.251183 | standard |
| 2.65673 | 0.0402106 | standard |
| 2.73251 | 0.0289084 | standard |
| 2.84511 | 0.622737 | standard |
| 2.87455 | 0.650087 | standard |
| 2.9233 | 1.0 | standard |
| 2.92711 | 0.995377 | standard |
| 3.06272 | 0.0482928 | standard |
| 3.1185 | 0.022323 | standard |
| 3.92532 | 0.377304 | standard |
| 3.98152 | 0.383471 | standard |
| 4.00045 | 0.373281 | standard |
| 4.05668 | 0.300187 | standard |
| 2.67844 | 0.0526315 | standard |
| 2.74715 | 0.038761 | standard |
| 2.84793 | 0.695547 | standard |
| 2.87857 | 0.730898 | standard |
| 2.91746 | 1.0 | standard |
| 2.92725 | 0.986215 | standard |
| 3.04805 | 0.0617306 | standard |
| 3.0978 | 0.0313861 | standard |
| 3.93147 | 0.424994 | standard |
| 3.98153 | 0.432363 | standard |
| 3.99965 | 0.421223 | standard |
| 4.04974 | 0.34591 | standard |
| 2.77275 | 0.140909 | standard |
| 2.80896 | 0.126807 | standard |
| 2.85756 | 0.724425 | standard |
| 2.89422 | 1.00003 | standard |
| 2.89925 | 0.965755 | standard |
| 2.9228 | 0.811993 | standard |
| 2.98461 | 0.152879 | standard |
| 3.0076 | 0.117038 | standard |
| 3.95973 | 0.469044 | standard |
| 3.98277 | 0.487033 | standard |
| 3.99585 | 0.476656 | standard |
| 4.01891 | 0.439322 | standard |
| 2.80181 | 0.284856 | standard |
| 2.82661 | 0.267618 | standard |
| 2.85825 | 0.926623 | standard |
| 2.88304 | 1.0 | standard |
| 2.90924 | 0.96322 | standard |
| 2.9239 | 0.993176 | standard |
| 2.96567 | 0.300876 | standard |
| 2.98033 | 0.26509 | standard |
| 3.96934 | 0.6473 | standard |
| 3.98403 | 0.66251 | standard |
| 3.9941 | 0.64351 | standard |
| 4.00879 | 0.619507 | standard |
| 2.8154 | 0.385372 | standard |
| 2.83424 | 0.366779 | standard |
| 2.85785 | 0.959332 | standard |
| 2.87669 | 0.983035 | standard |
| 2.91469 | 0.977145 | standard |
| 2.92539 | 1.00002 | standard |
| 2.95704 | 0.394383 | standard |
| 2.96784 | 0.368928 | standard |
| 3.9742 | 0.689196 | standard |
| 3.98491 | 0.717543 | standard |
| 3.99302 | 0.717232 | standard |
| 4.00384 | 0.688808 | standard |
| 2.82323 | 0.457391 | standard |
| 2.83841 | 0.444517 | standard |
| 2.85715 | 0.964497 | standard |
| 2.87233 | 0.980889 | standard |
| 2.91836 | 0.981941 | standard |
| 2.92678 | 1.00002 | standard |
| 2.95218 | 0.468041 | standard |
| 2.9607 | 0.448355 | standard |
| 3.97708 | 0.72923 | standard |
| 3.9856 | 0.757313 | standard |
| 3.99233 | 0.757653 | standard |
| 4.00076 | 0.729761 | standard |
| 2.82829 | 0.515633 | standard |
| 2.84099 | 0.5068 | standard |
| 2.85656 | 0.968812 | standard |
| 2.86925 | 0.980456 | standard |
| 2.92094 | 0.985597 | standard |
| 2.92797 | 1.00002 | standard |
| 2.9492 | 0.527651 | standard |
| 2.95623 | 0.512028 | standard |
| 3.97906 | 0.761941 | standard |
| 3.9861 | 0.79088 | standard |
| 3.99174 | 0.790863 | standard |
| 3.99878 | 0.761942 | standard |
| 2.83186 | 0.562997 | standard |
| 2.84277 | 0.557077 | standard |
| 2.85606 | 0.971942 | standard |
| 2.86697 | 0.980266 | standard |
| 2.92282 | 0.988259 | standard |
| 2.92886 | 1.00002 | standard |
| 2.94712 | 0.576377 | standard |
| 2.95306 | 0.563661 | standard |
| 3.98045 | 0.787812 | standard |
| 3.98649 | 0.817601 | standard |
| 3.99134 | 0.817589 | standard |
| 3.99739 | 0.787812 | standard |
| 2.83325 | 0.583404 | standard |
| 2.84347 | 0.578705 | standard |
| 2.85577 | 0.973184 | standard |
| 2.86598 | 0.980166 | standard |
| 2.92361 | 0.989298 | standard |
| 2.92926 | 1.00002 | standard |
| 2.94623 | 0.596704 | standard |
| 2.95187 | 0.585053 | standard |
| 3.98104 | 0.79964 | standard |
| 3.98659 | 0.828141 | standard |
| 3.99125 | 0.828132 | standard |
| 3.9968 | 0.79964 | standard |
| 2.83444 | 0.601162 | standard |
| 2.84396 | 0.597347 | standard |
| 2.85557 | 0.972667 | standard |
| 2.86519 | 0.978571 | standard |
| 2.92441 | 0.99026 | standard |
| 2.92956 | 1.00002 | standard |
| 2.94553 | 0.615006 | standard |
| 2.95078 | 0.604358 | standard |
| 3.98154 | 0.808216 | standard |
| 3.98679 | 0.838566 | standard |
| 3.99105 | 0.838558 | standard |
| 3.9963 | 0.808216 | standard |
| 2.83642 | 0.634138 | standard |
| 2.84495 | 0.631907 | standard |
| 2.85527 | 0.975418 | standard |
| 2.8637 | 0.979567 | standard |
| 2.9256 | 0.991672 | standard |
| 2.93025 | 1.00002 | standard |
| 2.94444 | 0.649576 | standard |
| 2.949 | 0.64051 | standard |
| 3.98233 | 0.826479 | standard |
| 3.98699 | 0.856256 | standard |
| 3.99085 | 0.856251 | standard |
| 3.99551 | 0.826479 | standard |
| 2.83722 | 0.648107 | standard |
| 2.84525 | 0.64644 | standard |
| 2.85507 | 0.975348 | standard |
| 2.8631 | 0.978794 | standard |
| 2.92609 | 0.992295 | standard |
| 2.93045 | 1.00002 | standard |
| 2.94395 | 0.663408 | standard |
| 2.9483 | 0.654972 | standard |
| 3.98263 | 0.833101 | standard |
| 3.98709 | 0.863223 | standard |
| 3.99075 | 0.864052 | standard |
| 3.99511 | 0.834282 | standard |
| 2.83791 | 0.66131 | standard |
| 2.84565 | 0.660331 | standard |
| 2.85487 | 0.976044 | standard |
| 2.86261 | 0.978765 | standard |
| 2.92659 | 0.992798 | standard |
| 2.93075 | 1.00002 | standard |
| 2.94355 | 0.676883 | standard |
| 2.94771 | 0.66898 | standard |
| 3.98302 | 0.841387 | standard |
| 3.98719 | 0.871839 | standard |
| 3.99065 | 0.871835 | standard |
| 3.99482 | 0.841387 | standard |
| 2.8392 | 0.68464 | standard |
| 2.84624 | 0.684656 | standard |
| 2.85458 | 0.976796 | standard |
| 2.86162 | 0.978398 | standard |
| 2.92738 | 0.993651 | standard |
| 2.93114 | 1.00002 | standard |
| 2.94275 | 0.70091 | standard |
| 2.94652 | 0.693917 | standard |
| 3.98352 | 0.853922 | standard |
| 3.98729 | 0.884296 | standard |
| 3.99045 | 0.88331 | standard |
| 3.99432 | 0.85253 | standard |
| 2.84109 | 0.722504 | standard |
| 2.84704 | 0.723689 | standard |
| 2.85418 | 0.977865 | standard |
| 2.86013 | 0.978031 | standard |
| 2.92867 | 0.994876 | standard |
| 2.93184 | 1.00002 | standard |
| 2.94166 | 0.738562 | standard |
| 2.94493 | 0.732827 | standard |
| 3.98431 | 0.872595 | standard |
| 3.98758 | 0.904317 | standard |
| 3.99026 | 0.905513 | standard |
| 3.99343 | 0.87429 | standard |