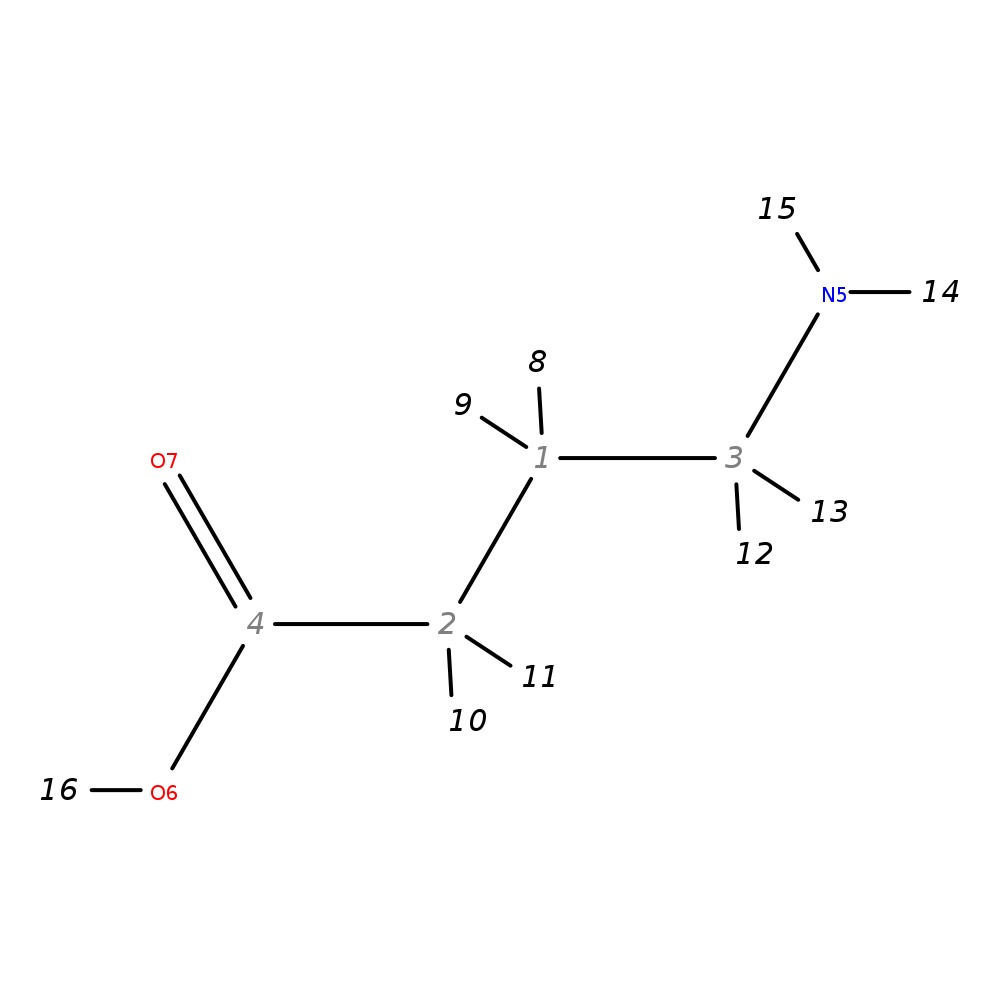
Gamma-Aminobutyric acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.13(MHz) | |
| RMSD of the fit | 0.01381 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C4H9NO2/c5-3-1-2-4(6)7/h1-3,5H2,(H,6,7) | |
| Note 1 | Oscar Li |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| gamma-Aminobutyric acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 1% |
Spin System Matrix
| 8 | 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|---|
| 8 | 1.893 | -17.296 | 7.194 | 8.413 | 6.335 | 8.905 |
| 9 | 0 | 1.893 | 6.758 | 7.811 | 9.543 | 5.644 |
| 10 | 0 | 0 | 2.286 | -11.578 | 0 | 0 |
| 11 | 0 | 0 | 0 | 2.286 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 2.997 | -12.394 |
| 13 | 0 | 0 | 0 | 0 | 0 | 2.997 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.86779 | 0.125669 | standard |
| 1.88032 | 0.451149 | standard |
| 1.89295 | 0.685426 | standard |
| 1.9056 | 0.498793 | standard |
| 1.91819 | 0.143029 | standard |
| 2.2737 | 0.545647 | standard |
| 2.28626 | 1.00022 | standard |
| 2.29881 | 0.472131 | standard |
| 2.98477 | 0.520504 | standard |
| 2.99754 | 0.874827 | standard |
| 3.01009 | 0.519019 | standard |
| 1.43517 | 0.0340221 | standard |
| 1.52662 | 0.0518682 | standard |
| 1.59932 | 0.0950901 | standard |
| 1.70724 | 0.190296 | standard |
| 1.84548 | 0.407379 | standard |
| 1.87542 | 0.380465 | standard |
| 1.99986 | 0.478694 | standard |
| 2.07725 | 0.47743 | standard |
| 2.15481 | 1.0 | standard |
| 2.28859 | 0.832725 | standard |
| 2.40447 | 0.187725 | standard |
| 2.45439 | 0.210773 | standard |
| 2.53475 | 0.128635 | standard |
| 2.839 | 0.492487 | standard |
| 2.91448 | 0.247103 | standard |
| 2.99857 | 0.78563 | standard |
| 3.18401 | 0.336952 | standard |
| 1.60061 | 0.0517625 | standard |
| 1.64577 | 0.0663671 | standard |
| 1.7194 | 0.168815 | standard |
| 1.76182 | 0.25394 | standard |
| 1.87434 | 0.554368 | standard |
| 1.99047 | 0.589074 | standard |
| 2.08446 | 0.292271 | standard |
| 2.10845 | 0.333614 | standard |
| 2.19078 | 0.837808 | standard |
| 2.28906 | 0.887245 | standard |
| 2.40359 | 0.256515 | standard |
| 2.44352 | 0.182273 | standard |
| 2.6386 | 0.0199161 | standard |
| 2.8867 | 0.641083 | standard |
| 3.00088 | 1.0 | standard |
| 3.12461 | 0.464115 | standard |
| 1.68004 | 0.0592591 | standard |
| 1.7064 | 0.070888 | standard |
| 1.77359 | 0.214306 | standard |
| 1.79347 | 0.277846 | standard |
| 1.88256 | 0.583212 | standard |
| 1.9721 | 0.583914 | standard |
| 2.04839 | 0.211638 | standard |
| 2.06737 | 0.225753 | standard |
| 2.20552 | 0.720749 | standard |
| 2.28882 | 0.84166 | standard |
| 2.3767 | 0.266149 | standard |
| 2.39943 | 0.219231 | standard |
| 2.65506 | 0.00860512 | standard |
| 2.91093 | 0.655988 | standard |
| 3.00141 | 1.0 | standard |
| 3.09374 | 0.483858 | standard |
| 1.70589 | 0.06288 | standard |
| 1.72671 | 0.0728181 | standard |
| 1.80447 | 0.290448 | standard |
| 1.8845 | 0.599712 | standard |
| 1.96433 | 0.556033 | standard |
| 2.03494 | 0.194682 | standard |
| 2.05109 | 0.203742 | standard |
| 2.21278 | 0.710965 | standard |
| 2.28898 | 0.82884 | standard |
| 2.36745 | 0.275694 | standard |
| 2.38499 | 0.239415 | standard |
| 2.9187 | 0.648261 | standard |
| 3.00112 | 1.00001 | standard |
| 3.08319 | 0.493223 | standard |
| 1.72634 | 0.066342 | standard |
| 1.74289 | 0.074865 | standard |
| 1.81327 | 0.301473 | standard |
| 1.88585 | 0.612085 | standard |
| 1.95853 | 0.562245 | standard |
| 2.02355 | 0.183255 | standard |
| 2.03705 | 0.188736 | standard |
| 2.2185 | 0.689336 | standard |
| 2.28854 | 0.851091 | standard |
| 2.35993 | 0.287774 | standard |
| 2.3735 | 0.258112 | standard |
| 2.92625 | 0.655589 | standard |
| 3.00078 | 1.0 | standard |
| 3.07468 | 0.501415 | standard |
| 1.8161 | 0.100245 | standard |
| 1.85303 | 0.40242 | standard |
| 1.89076 | 0.706389 | standard |
| 1.92829 | 0.569783 | standard |
| 1.96585 | 0.163909 | standard |
| 2.25038 | 0.642366 | standard |
| 2.28771 | 1.0 | standard |
| 2.32521 | 0.409177 | standard |
| 2.9603 | 0.619751 | standard |
| 2.9985 | 0.984212 | standard |
| 3.03587 | 0.543843 | standard |
| 1.8419 | 0.112332 | standard |
| 1.86685 | 0.424915 | standard |
| 1.89206 | 0.694456 | standard |
| 1.91725 | 0.533057 | standard |
| 1.94244 | 0.153894 | standard |
| 2.2618 | 0.589981 | standard |
| 2.28686 | 1.00006 | standard |
| 2.31194 | 0.441847 | standard |
| 2.97243 | 0.563236 | standard |
| 2.99789 | 0.916429 | standard |
| 3.02297 | 0.524516 | standard |
| 1.85489 | 0.118154 | standard |
| 1.87364 | 0.440806 | standard |
| 1.89259 | 0.689361 | standard |
| 1.91151 | 0.510993 | standard |
| 1.93042 | 0.149363 | standard |
| 2.26769 | 0.564741 | standard |
| 2.28651 | 1.00012 | standard |
| 2.30533 | 0.456853 | standard |
| 2.97857 | 0.537309 | standard |
| 2.99767 | 0.893992 | standard |
| 3.01653 | 0.522131 | standard |
| 1.86264 | 0.124317 | standard |
| 1.87768 | 0.444895 | standard |
| 1.89282 | 0.685197 | standard |
| 1.90797 | 0.501012 | standard |
| 1.92309 | 0.144494 | standard |
| 2.2713 | 0.553194 | standard |
| 2.28635 | 1.00007 | standard |
| 2.30145 | 0.468965 | standard |
| 2.98231 | 0.524465 | standard |
| 2.99757 | 0.881587 | standard |
| 3.01268 | 0.523288 | standard |
| 1.86779 | 0.125672 | standard |
| 1.88032 | 0.451162 | standard |
| 1.89295 | 0.685445 | standard |
| 1.9056 | 0.498806 | standard |
| 1.91819 | 0.143036 | standard |
| 2.2737 | 0.545657 | standard |
| 2.28626 | 1.00022 | standard |
| 2.29881 | 0.472132 | standard |
| 2.98477 | 0.520502 | standard |
| 2.99754 | 0.874827 | standard |
| 3.01011 | 0.519553 | standard |
| 1.87146 | 0.127877 | standard |
| 1.88222 | 0.455694 | standard |
| 1.89303 | 0.681243 | standard |
| 1.90387 | 0.488646 | standard |
| 1.91467 | 0.141532 | standard |
| 2.27543 | 0.536895 | standard |
| 2.2862 | 1.00072 | standard |
| 2.29694 | 0.483235 | standard |
| 2.98658 | 0.518515 | standard |
| 2.99749 | 0.871568 | standard |
| 3.00828 | 0.517096 | standard |
| 1.87289 | 0.12918 | standard |
| 1.88297 | 0.462396 | standard |
| 1.89307 | 0.686555 | standard |
| 1.90317 | 0.485628 | standard |
| 1.91328 | 0.140668 | standard |
| 2.27613 | 0.533961 | standard |
| 2.28619 | 1.0011 | standard |
| 2.29625 | 0.484593 | standard |
| 2.98729 | 0.519453 | standard |
| 2.99748 | 0.872621 | standard |
| 3.00756 | 0.518501 | standard |
| 1.87418 | 0.129195 | standard |
| 1.88363 | 0.4628 | standard |
| 1.89308 | 0.683345 | standard |
| 1.90258 | 0.488986 | standard |
| 1.91204 | 0.141222 | standard |
| 2.27677 | 0.533142 | standard |
| 2.28617 | 1.00089 | standard |
| 2.2956 | 0.490103 | standard |
| 2.98794 | 0.517851 | standard |
| 2.99746 | 0.871269 | standard |
| 3.00697 | 0.517563 | standard |
| 1.87631 | 0.131215 | standard |
| 1.88472 | 0.463008 | standard |
| 1.89314 | 0.681681 | standard |
| 1.90157 | 0.484466 | standard |
| 1.90996 | 0.138692 | standard |
| 2.27776 | 0.526223 | standard |
| 2.28614 | 1.0 | standard |
| 2.29451 | 0.490313 | standard |
| 2.98898 | 0.516736 | standard |
| 2.9974 | 0.868906 | standard |
| 3.00588 | 0.515116 | standard |
| 1.87721 | 0.131475 | standard |
| 1.88515 | 0.463941 | standard |
| 1.89313 | 0.681054 | standard |
| 1.90113 | 0.482465 | standard |
| 1.90912 | 0.139763 | standard |
| 2.27826 | 0.524143 | standard |
| 2.28614 | 1.0 | standard |
| 2.29407 | 0.498078 | standard |
| 2.98943 | 0.515793 | standard |
| 2.9974 | 0.872016 | standard |
| 3.00538 | 0.515773 | standard |
| 1.878 | 0.132159 | standard |
| 1.8856 | 0.46878 | standard |
| 1.89315 | 0.688843 | standard |
| 1.90074 | 0.481131 | standard |
| 1.90833 | 0.138949 | standard |
| 2.2786 | 0.522236 | standard |
| 2.28614 | 1.0 | standard |
| 2.29367 | 0.501492 | standard |
| 2.98982 | 0.516281 | standard |
| 2.9974 | 0.864672 | standard |
| 3.00499 | 0.51616 | standard |
| 1.87944 | 0.134056 | standard |
| 1.88631 | 0.470972 | standard |
| 1.89317 | 0.687265 | standard |
| 1.90008 | 0.47888 | standard |
| 1.90694 | 0.137758 | standard |
| 2.27925 | 0.517233 | standard |
| 2.28612 | 1.00046 | standard |
| 2.29298 | 0.503133 | standard |
| 2.99047 | 0.517427 | standard |
| 2.9974 | 0.864621 | standard |
| 3.00429 | 0.515573 | standard |
| 1.88157 | 0.13329 | standard |
| 1.88738 | 0.473404 | standard |
| 1.8932 | 0.68613 | standard |
| 1.89904 | 0.480317 | standard |
| 1.90486 | 0.135225 | standard |
| 2.28029 | 0.51692 | standard |
| 2.28609 | 1.00435 | standard |
| 2.29194 | 0.508633 | standard |
| 2.99156 | 0.517518 | standard |
| 2.9974 | 0.859404 | standard |
| 3.00325 | 0.517376 | standard |