3-Methylsalicylaldehyde
Simulation outputs:
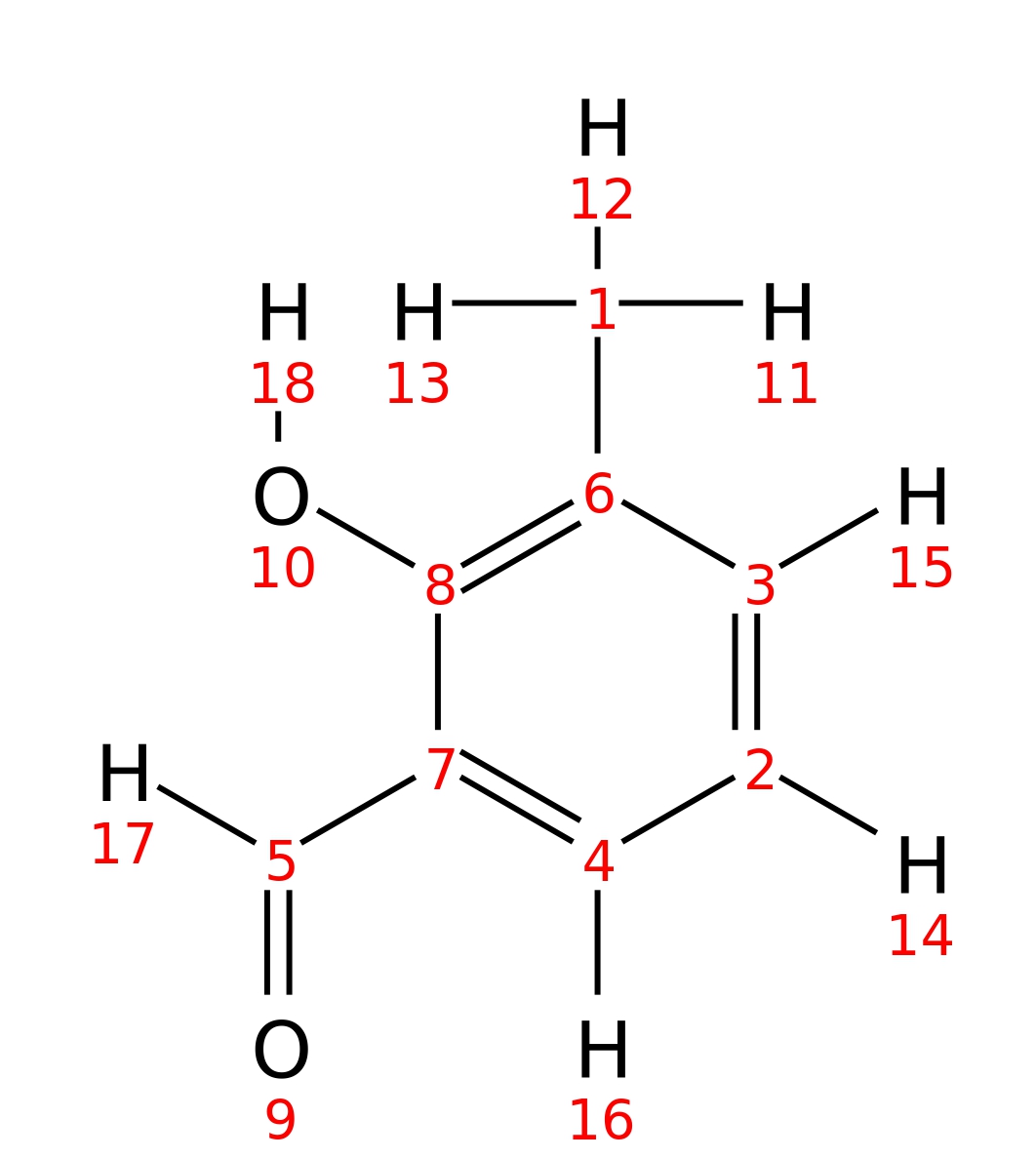
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01530 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H8O2/c1-6-3-2-4-7(5-9)8(6)10/h2-5,10H,1H3 | |
| Note 1 | 15?16 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-Methylsalicylaldehyde | Solute | 100mM |
| CDCl3 | Solvent | 100% |
| TMS | Reference | 0.05mM |
Spin System Matrix
| 11 | 12 | 13 | 15 | 14 | 16 | 17 | |
|---|---|---|---|---|---|---|---|
| 11 | 2.275 | -14.9 | -14.9 | 0.614 | 0 | 0 | 0 |
| 12 | 0 | 2.275 | -14.9 | 0.614 | 0 | 0 | 0 |
| 13 | 0 | 0 | 2.275 | 0.614 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 7.395 | 7.531 | 1.394 | 0 |
| 14 | 0 | 0 | 0 | 0 | 6.931 | 7.531 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 7.403 | 0.532 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 9.879 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.27461 | 1.00001 | standard |
| 6.91616 | 0.10847 | standard |
| 6.93123 | 0.222464 | standard |
| 6.94631 | 0.117829 | standard |
| 7.38862 | 0.118709 | standard |
| 7.38934 | 0.116472 | standard |
| 7.39422 | 0.146295 | standard |
| 7.39629 | 0.0900191 | standard |
| 7.40375 | 0.114325 | standard |
| 7.40917 | 0.134404 | standard |
| 9.87926 | 0.352352 | standard |
| -0.994301 | 1.125e-06 | standard |
| 2.2746 | 1.0 | standard |
| 6.71329 | 0.0469503 | standard |
| 6.8584 | 0.0559491 | standard |
| 6.93295 | 0.0847583 | standard |
| 7.07709 | 0.199091 | standard |
| 7.32573 | 0.356438 | standard |
| 7.48101 | 0.111994 | standard |
| 7.53803 | 0.0926071 | standard |
| 9.87925 | 0.349088 | standard |
| 2.27467 | 1.0 | standard |
| 6.7915 | 0.0610005 | standard |
| 6.90077 | 0.0832452 | standard |
| 6.92936 | 0.0971142 | standard |
| 7.03835 | 0.169343 | standard |
| 7.34623 | 0.336582 | standard |
| 7.46318 | 0.15102 | standard |
| 9.87928 | 0.348898 | standard |
| 2.2747 | 1.00001 | standard |
| 6.82888 | 0.0693031 | standard |
| 6.91435 | 0.0995576 | standard |
| 6.92995 | 0.107772 | standard |
| 7.01533 | 0.151332 | standard |
| 7.35791 | 0.32329 | standard |
| 7.4495 | 0.195462 | standard |
| 9.87927 | 0.348241 | standard |
| 2.2747 | 1.00001 | standard |
| 6.84101 | 0.0729231 | standard |
| 6.91817 | 0.106467 | standard |
| 6.93 | 0.112459 | standard |
| 7.00708 | 0.145856 | standard |
| 7.36203 | 0.317831 | standard |
| 7.44391 | 0.2138 | standard |
| 9.87928 | 0.348066 | standard |
| 2.27471 | 1.00001 | standard |
| 6.85061 | 0.0757279 | standard |
| 6.92089 | 0.112275 | standard |
| 6.93007 | 0.117191 | standard |
| 7.00029 | 0.141662 | standard |
| 7.36541 | 0.313707 | standard |
| 7.4393 | 0.225214 | standard |
| 9.87928 | 0.348586 | standard |
| 2.27473 | 1.0 | standard |
| 6.89235 | 0.091962 | standard |
| 6.92998 | 0.17864 | standard |
| 6.96753 | 0.126231 | standard |
| 7.38197 | 0.258904 | standard |
| 7.41905 | 0.237944 | standard |
| 9.87929 | 0.35167 | standard |
| 2.27473 | 1.0 | standard |
| 6.90574 | 0.100097 | standard |
| 6.9308 | 0.208894 | standard |
| 6.95586 | 0.123347 | standard |
| 7.38835 | 0.19985 | standard |
| 7.41299 | 0.187086 | standard |
| 9.87927 | 0.355161 | standard |
| 2.27458 | 1.0001 | standard |
| 6.91229 | 0.104688 | standard |
| 6.9311 | 0.219592 | standard |
| 6.94988 | 0.12202 | standard |
| 7.38786 | 0.133523 | standard |
| 7.39208 | 0.165244 | standard |
| 7.40597 | 0.118716 | standard |
| 7.40701 | 0.120528 | standard |
| 7.41059 | 0.146484 | standard |
| 9.87927 | 0.359661 | standard |
| 2.27462 | 1.0 | standard |
| 2.27479 | 0.99988 | standard |
| 6.91616 | 0.108603 | standard |
| 6.93123 | 0.221937 | standard |
| 6.94631 | 0.117819 | standard |
| 7.38862 | 0.119086 | standard |
| 7.38933 | 0.116662 | standard |
| 7.39421 | 0.146759 | standard |
| 7.39629 | 0.0901228 | standard |
| 7.40376 | 0.114037 | standard |
| 7.40917 | 0.134749 | standard |
| 9.87926 | 0.352853 | standard |
| 2.27462 | 0.999952 | standard |
| 2.27485 | 1.00001 | standard |
| 6.91874 | 0.111067 | standard |
| 6.93128 | 0.225649 | standard |
| 6.94383 | 0.116574 | standard |
| 7.38973 | 0.114609 | standard |
| 7.39562 | 0.142348 | standard |
| 7.39753 | 0.0978691 | standard |
| 7.40229 | 0.114763 | standard |
| 7.4081 | 0.13199 | standard |
| 7.40988 | 0.08745 | standard |
| 9.87927 | 0.356611 | standard |
| 2.27482 | 1.00002 | standard |
| 6.92062 | 0.11251 | standard |
| 6.93135 | 0.226448 | standard |
| 6.94214 | 0.115452 | standard |
| 7.39047 | 0.111516 | standard |
| 7.39673 | 0.137009 | standard |
| 7.39891 | 0.126715 | standard |
| 7.4003 | 0.111234 | standard |
| 7.40122 | 0.116492 | standard |
| 7.40742 | 0.124866 | standard |
| 7.40878 | 0.0906422 | standard |
| 9.87929 | 0.355226 | standard |
| 2.27466 | 1.00008 | standard |
| 2.2748 | 1.00007 | standard |
| 6.92131 | 0.112762 | standard |
| 6.93138 | 0.225274 | standard |
| 6.94145 | 0.114876 | standard |
| 7.39089 | 0.109136 | standard |
| 7.39715 | 0.137107 | standard |
| 7.39898 | 0.146261 | standard |
| 7.40081 | 0.120703 | standard |
| 7.40702 | 0.124441 | standard |
| 7.40843 | 0.0908166 | standard |
| 9.8793 | 0.355037 | standard |
| 2.27456 | 1.00025 | standard |
| 2.2749 | 0.999761 | standard |
| 6.92201 | 0.116693 | standard |
| 6.93138 | 0.231675 | standard |
| 6.94075 | 0.118041 | standard |
| 7.39111 | 0.112252 | standard |
| 7.39759 | 0.148548 | standard |
| 7.39902 | 0.163614 | standard |
| 7.40044 | 0.130079 | standard |
| 7.40672 | 0.129943 | standard |
| 7.40803 | 0.0933331 | standard |
| 9.87927 | 0.362265 | standard |
| 2.27455 | 1.00014 | standard |
| 2.27481 | 1.00007 | standard |
| 6.923 | 0.116552 | standard |
| 6.93138 | 0.229485 | standard |
| 6.93976 | 0.11664 | standard |
| 7.39151 | 0.112211 | standard |
| 7.39815 | 0.176377 | standard |
| 7.39922 | 0.184453 | standard |
| 7.4063 | 0.126406 | standard |
| 7.40743 | 0.0961671 | standard |
| 9.87925 | 0.358258 | standard |
| 2.27473 | 1.0 | standard |
| 6.9235 | 0.110616 | standard |
| 6.93143 | 0.220769 | standard |
| 6.93937 | 0.111106 | standard |
| 7.39172 | 0.104111 | standard |
| 7.3984 | 0.185773 | standard |
| 7.39955 | 0.19001 | standard |
| 7.40607 | 0.120091 | standard |
| 7.40731 | 0.092501 | standard |
| 9.87925 | 0.333473 | standard |
| 2.27468 | 1.0008 | standard |
| 2.27479 | 1.0008 | standard |
| 6.92389 | 0.113744 | standard |
| 6.93143 | 0.226592 | standard |
| 6.93897 | 0.113557 | standard |
| 7.39194 | 0.106938 | standard |
| 7.39862 | 0.201206 | standard |
| 7.39947 | 0.195461 | standard |
| 7.40602 | 0.116094 | standard |
| 7.40689 | 0.0928773 | standard |
| 9.8793 | 0.361174 | standard |
| 2.27458 | 1.00291 | standard |
| 2.27489 | 1.00294 | standard |
| 6.92459 | 0.11933 | standard |
| 6.93143 | 0.236794 | standard |
| 6.93827 | 0.118813 | standard |
| 7.39222 | 0.111497 | standard |
| 7.3988 | 0.227076 | standard |
| 7.40561 | 0.126949 | standard |
| 7.40655 | 0.0986605 | standard |
| 9.8793 | 0.363975 | standard |
| 2.27457 | 1.00251 | standard |
| 2.27479 | 1.00261 | standard |
| 6.92568 | 0.118394 | standard |
| 6.93143 | 0.234823 | standard |
| 6.93718 | 0.118301 | standard |
| 7.39271 | 0.109292 | standard |
| 7.39909 | 0.196378 | standard |
| 7.40016 | 0.113473 | standard |
| 7.40513 | 0.120456 | standard |
| 7.40599 | 0.0995357 | standard |
| 9.87925 | 0.369014 | standard |