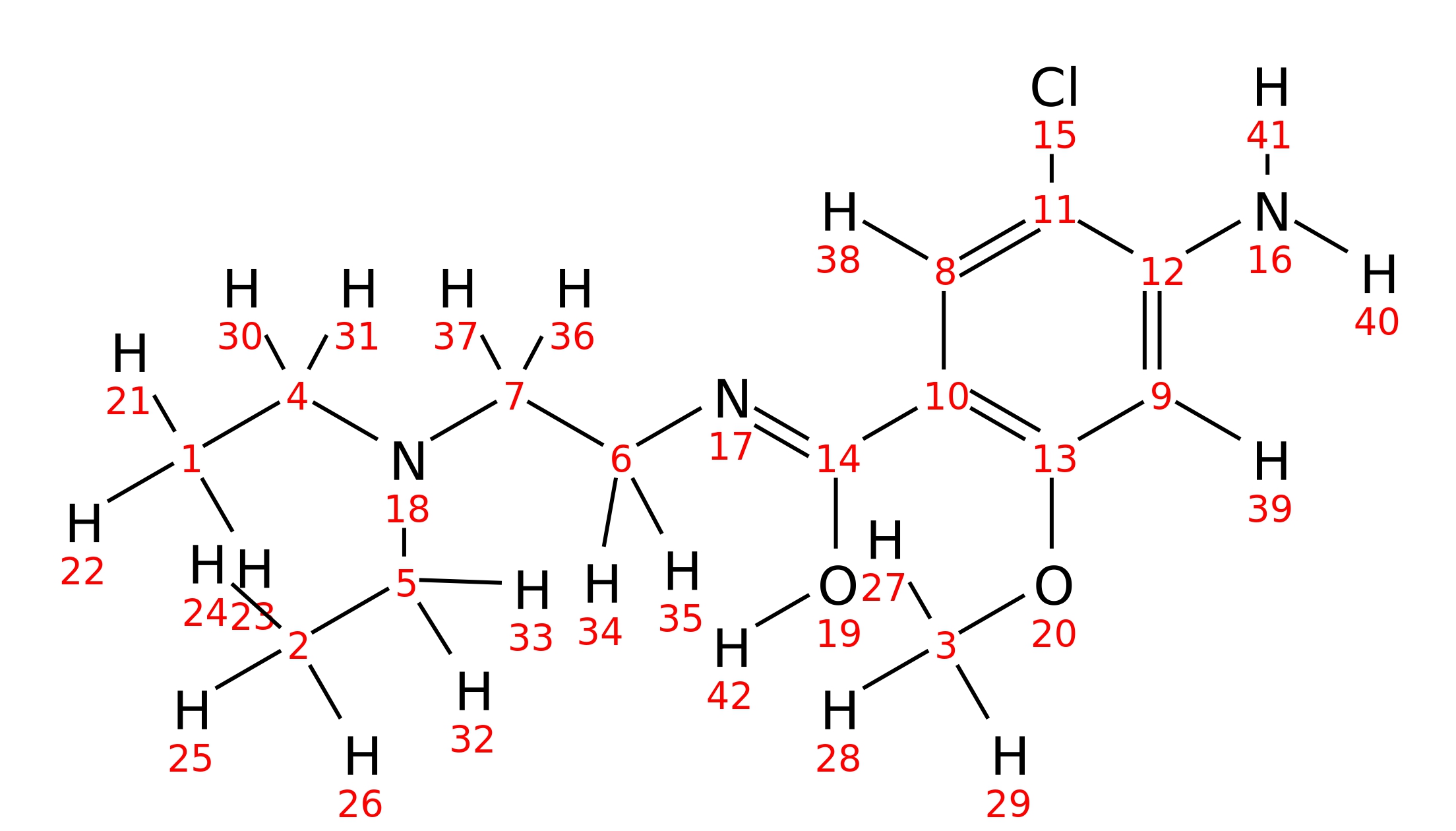
Metoclopramide
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.02049 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C14H22ClN3O2/c1-4-18(5-2)7-6-17-14(19)10-8-11(15)12(16)9-13(10)20-3/h8-9H,4-7,16H2,1-3H3,(H,17,19) | |
| Note 1 | ||
| Note 2 | 38 @7.9 is multiplet (35/37 Cl isotope shifts?) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Metoclopramide | Solute | 100mM |
| methanol | Solvent | 100% |
| TMS | Reference | 0.05mM |
Spin System Matrix
| 21 | 22 | 23 | 30 | 31 | 32 | 33 | 24 | 25 | 26 | 36 | 37 | 34 | 35 | 38 | 39 | 27 | 28 | 29 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21 | 1.472 | -12.5 | -12.5 | 7.258 | 7.258 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 1.472 | -12.5 | 7.258 | 7.258 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 1.472 | 7.258 | 7.258 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 30 | 0 | 0 | 0 | 3.397 | -12.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 31 | 0 | 0 | 0 | 0 | 3.397 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 32 | 0 | 0 | 0 | 0 | 0 | 3.397 | -12.4 | 7.258 | 7.258 | 7.258 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 33 | 0 | 0 | 0 | 0 | 0 | 0 | 3.397 | 7.258 | 7.258 | 7.258 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.472 | -12.5 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.472 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.472 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 36 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.454 | -12.4 | 6.074 | 6.074 | 0 | 0 | 0 | 0 | 0 |
| 37 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.454 | 6.074 | 6.074 | 0 | 0 | 0 | 0 | 0 |
| 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.885 | -12.4 | 0 | 0 | 0 | 0 | 0 |
| 35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.885 | 0 | 0 | 0 | 0 | 0 |
| 38 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.973 | 0.1 | 0 | 0 | 0 |
| 39 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.67 | 0 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.081 | -11.5 | -11.5 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.081 | -11.5 |
| 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.081 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.45745 | 0.511476 | standard |
| 1.47197 | 1.0006 | standard |
| 1.48649 | 0.511476 | standard |
| 3.37552 | 0.175769 | standard |
| 3.38999 | 0.504343 | standard |
| 3.40451 | 0.504926 | standard |
| 3.41903 | 0.177871 | standard |
| 3.44168 | 0.16938 | standard |
| 3.45381 | 0.33578 | standard |
| 3.46594 | 0.180196 | standard |
| 3.87329 | 0.180028 | standard |
| 3.88539 | 0.333559 | standard |
| 3.8975 | 0.165524 | standard |
| 4.08131 | 0.983641 | standard |
| 6.66963 | 0.325474 | standard |
| 7.97323 | 0.325584 | standard |
| 1.28498 | 0.34461 | standard |
| 1.46473 | 0.853524 | standard |
| 1.64294 | 0.51109 | standard |
| 3.14048 | 0.190462 | standard |
| 3.31454 | 0.502486 | standard |
| 3.494 | 0.461307 | standard |
| 3.56143 | 0.311082 | standard |
| 3.6861 | 0.153623 | standard |
| 3.76258 | 0.265827 | standard |
| 3.88892 | 0.25006 | standard |
| 3.96819 | 0.128448 | standard |
| 4.08133 | 1.0 | standard |
| 6.66963 | 0.315016 | standard |
| 7.97323 | 0.314792 | standard |
| 1.34806 | 0.416808 | standard |
| 1.46837 | 0.953733 | standard |
| 1.58862 | 0.538039 | standard |
| 3.22226 | 0.197827 | standard |
| 3.34179 | 0.559181 | standard |
| 3.45853 | 0.635081 | standard |
| 3.54275 | 0.263498 | standard |
| 3.57849 | 0.182605 | standard |
| 3.79744 | 0.225382 | standard |
| 3.88854 | 0.254413 | standard |
| 3.98641 | 0.11339 | standard |
| 4.01038 | 0.123636 | standard |
| 4.08129 | 1.0 | standard |
| 6.66963 | 0.328942 | standard |
| 7.97323 | 0.328912 | standard |
| 1.37952 | 0.449364 | standard |
| 1.46993 | 0.981816 | standard |
| 1.56039 | 0.539643 | standard |
| 3.26467 | 0.196441 | standard |
| 3.35567 | 0.554584 | standard |
| 3.44633 | 0.71109 | standard |
| 3.52774 | 0.306204 | standard |
| 3.81696 | 0.212425 | standard |
| 3.88859 | 0.268779 | standard |
| 3.96288 | 0.110391 | standard |
| 4.08131 | 1.0 | standard |
| 6.66963 | 0.330731 | standard |
| 7.97323 | 0.330302 | standard |
| 1.38996 | 0.457925 | standard |
| 1.47036 | 0.989856 | standard |
| 1.55081 | 0.5397 | standard |
| 3.27902 | 0.195262 | standard |
| 3.35967 | 0.538109 | standard |
| 3.44148 | 0.680307 | standard |
| 3.51799 | 0.347932 | standard |
| 3.8238 | 0.208288 | standard |
| 3.88851 | 0.278115 | standard |
| 3.95539 | 0.114848 | standard |
| 4.08131 | 1.0 | standard |
| 6.66963 | 0.330709 | standard |
| 7.97323 | 0.330844 | standard |
| 1.39827 | 0.467453 | standard |
| 1.47068 | 0.994971 | standard |
| 1.5431 | 0.5404 | standard |
| 3.29061 | 0.194705 | standard |
| 3.36305 | 0.528304 | standard |
| 3.437 | 0.606887 | standard |
| 3.50878 | 0.35664 | standard |
| 3.82937 | 0.206044 | standard |
| 3.88834 | 0.286057 | standard |
| 3.94875 | 0.11949 | standard |
| 4.08131 | 1.0 | standard |
| 6.66963 | 0.331122 | standard |
| 7.97323 | 0.330938 | standard |
| 1.43545 | 0.49942 | standard |
| 1.47168 | 1.00005 | standard |
| 1.50793 | 0.521487 | standard |
| 3.34333 | 0.181264 | standard |
| 3.37956 | 0.510163 | standard |
| 3.41609 | 0.55625 | standard |
| 3.45261 | 0.488728 | standard |
| 3.48315 | 0.199586 | standard |
| 3.856 | 0.192215 | standard |
| 3.88624 | 0.321888 | standard |
| 3.91653 | 0.146946 | standard |
| 4.08131 | 0.988469 | standard |
| 6.66963 | 0.328447 | standard |
| 7.97323 | 0.327701 | standard |
| 1.44774 | 0.507665 | standard |
| 1.47187 | 1.00021 | standard |
| 1.4961 | 0.512382 | standard |
| 3.36115 | 0.176169 | standard |
| 3.38533 | 0.502553 | standard |
| 3.40949 | 0.50938 | standard |
| 3.43352 | 0.330772 | standard |
| 3.4535 | 0.338436 | standard |
| 3.47371 | 0.189511 | standard |
| 3.86546 | 0.186618 | standard |
| 3.88569 | 0.329173 | standard |
| 3.90591 | 0.156494 | standard |
| 4.08131 | 0.98425 | standard |
| 6.66963 | 0.326702 | standard |
| 7.97323 | 0.326945 | standard |
| 1.45385 | 0.511053 | standard |
| 1.47197 | 1.00038 | standard |
| 1.49008 | 0.511527 | standard |
| 3.37012 | 0.175809 | standard |
| 3.38823 | 0.502351 | standard |
| 3.40636 | 0.508247 | standard |
| 3.42449 | 0.183258 | standard |
| 3.43854 | 0.17216 | standard |
| 3.45371 | 0.335716 | standard |
| 3.46886 | 0.18491 | standard |
| 3.87032 | 0.183112 | standard |
| 3.88549 | 0.331674 | standard |
| 3.90061 | 0.163259 | standard |
| 4.08131 | 0.983423 | standard |
| 6.66963 | 0.325603 | standard |
| 7.97323 | 0.325756 | standard |
| 1.45747 | 0.510699 | standard |
| 1.47197 | 1.0006 | standard |
| 1.48649 | 0.510381 | standard |
| 3.37552 | 0.175741 | standard |
| 3.39 | 0.503783 | standard |
| 3.40451 | 0.504937 | standard |
| 3.41903 | 0.177863 | standard |
| 3.44168 | 0.169124 | standard |
| 3.45381 | 0.335721 | standard |
| 3.46594 | 0.180315 | standard |
| 3.87329 | 0.179726 | standard |
| 3.88539 | 0.333285 | standard |
| 3.8975 | 0.165726 | standard |
| 4.08131 | 0.983237 | standard |
| 6.66963 | 0.325398 | standard |
| 7.97323 | 0.325589 | standard |
| 1.45993 | 0.510643 | standard |
| 1.47202 | 1.0 | standard |
| 1.48411 | 0.510643 | standard |
| 3.37914 | 0.175675 | standard |
| 3.39121 | 0.503846 | standard |
| 3.40327 | 0.502223 | standard |
| 3.41536 | 0.176227 | standard |
| 3.44376 | 0.171435 | standard |
| 3.45386 | 0.335527 | standard |
| 3.46396 | 0.177072 | standard |
| 3.87522 | 0.176539 | standard |
| 3.88532 | 0.333821 | standard |
| 3.89542 | 0.169657 | standard |
| 4.08131 | 0.982542 | standard |
| 6.66963 | 0.326651 | standard |
| 7.97323 | 0.326561 | standard |
| 1.46163 | 0.511034 | standard |
| 1.47202 | 1.0 | standard |
| 1.4824 | 0.511033 | standard |
| 3.38171 | 0.175612 | standard |
| 3.39204 | 0.503753 | standard |
| 3.40241 | 0.503919 | standard |
| 3.41273 | 0.176193 | standard |
| 3.44524 | 0.171831 | standard |
| 3.45391 | 0.336177 | standard |
| 3.46257 | 0.175948 | standard |
| 3.87665 | 0.176139 | standard |
| 3.88532 | 0.333686 | standard |
| 3.89393 | 0.171024 | standard |
| 4.08131 | 0.982264 | standard |
| 6.66963 | 0.326325 | standard |
| 7.97323 | 0.326565 | standard |
| 1.46233 | 0.511431 | standard |
| 1.47202 | 1.0 | standard |
| 1.48171 | 0.511431 | standard |
| 3.3827 | 0.175625 | standard |
| 3.39238 | 0.504329 | standard |
| 3.40205 | 0.504224 | standard |
| 3.41174 | 0.17605 | standard |
| 3.44584 | 0.172595 | standard |
| 3.45391 | 0.336233 | standard |
| 3.46198 | 0.174866 | standard |
| 3.8772 | 0.174048 | standard |
| 3.88529 | 0.334689 | standard |
| 3.89339 | 0.172131 | standard |
| 4.08131 | 0.983339 | standard |
| 6.66963 | 0.326456 | standard |
| 7.97323 | 0.326731 | standard |
| 1.46297 | 0.510988 | standard |
| 1.47202 | 1.0 | standard |
| 1.48111 | 0.51079 | standard |
| 3.3836 | 0.175362 | standard |
| 3.39268 | 0.503736 | standard |
| 3.40172 | 0.503582 | standard |
| 3.4108 | 0.17599 | standard |
| 3.44633 | 0.172361 | standard |
| 3.45391 | 0.335461 | standard |
| 3.46153 | 0.174841 | standard |
| 3.87769 | 0.173773 | standard |
| 3.88527 | 0.334296 | standard |
| 3.89284 | 0.171452 | standard |
| 4.08131 | 0.982964 | standard |
| 6.66963 | 0.326438 | standard |
| 7.97323 | 0.326737 | standard |
| 1.46396 | 0.510759 | standard |
| 1.47202 | 1.0 | standard |
| 1.48007 | 0.510759 | standard |
| 3.38513 | 0.175655 | standard |
| 3.39318 | 0.503542 | standard |
| 3.40122 | 0.503283 | standard |
| 3.40926 | 0.175571 | standard |
| 3.44717 | 0.173126 | standard |
| 3.45391 | 0.33533 | standard |
| 3.46064 | 0.174351 | standard |
| 3.87854 | 0.173751 | standard |
| 3.88527 | 0.334667 | standard |
| 3.892 | 0.17265 | standard |
| 4.08131 | 0.982602 | standard |
| 6.66963 | 0.326366 | standard |
| 7.97323 | 0.326469 | standard |
| 1.46439 | 0.511849 | standard |
| 1.47202 | 1.0 | standard |
| 1.47965 | 0.512442 | standard |
| 3.38573 | 0.175899 | standard |
| 3.39337 | 0.504361 | standard |
| 3.40101 | 0.503253 | standard |
| 3.40867 | 0.175677 | standard |
| 3.44757 | 0.173297 | standard |
| 3.45391 | 0.335663 | standard |
| 3.46035 | 0.174157 | standard |
| 3.87888 | 0.174221 | standard |
| 3.88527 | 0.335341 | standard |
| 3.89161 | 0.172748 | standard |
| 4.08131 | 0.983017 | standard |
| 6.66963 | 0.326388 | standard |
| 7.97323 | 0.326755 | standard |
| 1.46478 | 0.511562 | standard |
| 1.47202 | 1.0 | standard |
| 1.47928 | 0.508798 | standard |
| 3.38632 | 0.175627 | standard |
| 3.39356 | 0.503963 | standard |
| 3.40083 | 0.501477 | standard |
| 3.40808 | 0.175326 | standard |
| 3.44787 | 0.173466 | standard |
| 3.45393 | 0.335167 | standard |
| 3.46005 | 0.173073 | standard |
| 3.87923 | 0.173032 | standard |
| 3.88527 | 0.334191 | standard |
| 3.89131 | 0.17287 | standard |
| 4.08131 | 0.981561 | standard |
| 6.66963 | 0.326087 | standard |
| 7.97323 | 0.325832 | standard |
| 1.46545 | 0.511186 | standard |
| 1.47202 | 1.0 | standard |
| 1.47863 | 0.508828 | standard |
| 3.38731 | 0.175859 | standard |
| 3.39389 | 0.503325 | standard |
| 3.4005 | 0.504669 | standard |
| 3.40708 | 0.175437 | standard |
| 3.44846 | 0.173468 | standard |
| 3.45396 | 0.336221 | standard |
| 3.45945 | 0.1735 | standard |
| 3.87972 | 0.172899 | standard |
| 3.88527 | 0.334638 | standard |
| 3.89077 | 0.173669 | standard |
| 4.08131 | 0.982613 | standard |
| 6.66963 | 0.325888 | standard |
| 7.97323 | 0.326307 | standard |
| 1.46647 | 0.511683 | standard |
| 1.47202 | 1.0 | standard |
| 1.47764 | 0.509662 | standard |
| 3.3888 | 0.175512 | standard |
| 3.39438 | 0.501997 | standard |
| 3.4 | 0.503894 | standard |
| 3.40555 | 0.175708 | standard |
| 3.4493 | 0.173741 | standard |
| 3.45396 | 0.336186 | standard |
| 3.45861 | 0.173868 | standard |
| 3.88057 | 0.173423 | standard |
| 3.88522 | 0.335652 | standard |
| 3.88992 | 0.172556 | standard |
| 4.08131 | 0.98247 | standard |
| 6.66963 | 0.325643 | standard |
| 7.97323 | 0.325385 | standard |