Trimethyl Phosphate
Simulation outputs:
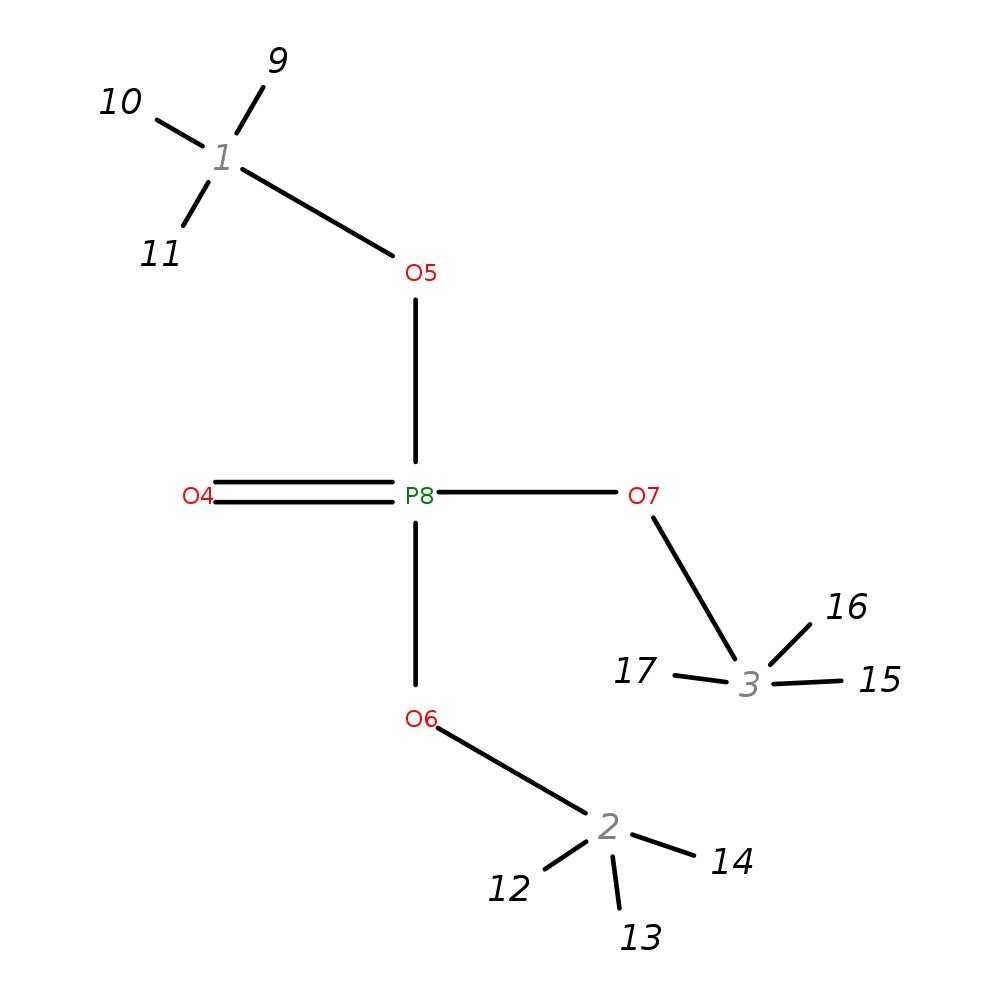
|
Parameter | Value | |||||||||||||||||||
| Field strength | 499.84(MHz) | ||||||||||||||||||||
| RMSD of the fit | 0.00736 | ||||||||||||||||||||
| Temperature | 298 K | ||||||||||||||||||||
| pH | 7.4 | ||||||||||||||||||||
| InChI | InChI=1S/C3H9O4P/c1-5-8(4,6-2)7-3/h1-3H3 | ||||||||||||||||||||
| Note 1 | |||||||||||||||||||||
| Note 2 | Non-hydrogen additional coupling; | ||||||||||||||||||||
| Note 3 | spin name(s):"9,10,11,12,13,14,15,16,17", coupling of "11.095923" with "P8" | ||||||||||||||||||||
| Additional coupling constants |
|
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Trimethyl Phosphate | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|---|---|---|
| 9 | 3.81 | -12.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 3.81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 3.81 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 3.81 | 0 | -12.4 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 3.81 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 3.81 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 3.81 | -12.4 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.81 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.81 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.79874 | 1.00003 | standard |
| 3.82094 | 1.00003 | standard |
| 3.67116 | 1.0 | standard |
| 3.94852 | 1.0 | standard |
| 3.71738 | 1.0 | standard |
| 3.90229 | 1.0 | standard |
| 3.7405 | 1.0 | standard |
| 3.87918 | 1.0 | standard |
| 3.7482 | 1.00001 | standard |
| 3.87148 | 1.00001 | standard |
| 3.75437 | 1.00001 | standard |
| 3.86531 | 1.00001 | standard |
| 3.7821 | 1.00005 | standard |
| 3.83758 | 1.00005 | standard |
| 3.79135 | 1.00016 | standard |
| 3.82833 | 1.00016 | standard |
| 3.79597 | 1.00005 | standard |
| 3.82371 | 1.00005 | standard |
| 3.79874 | 1.00005 | standard |
| 3.82093 | 1.00005 | standard |
| 3.80059 | 1.00016 | standard |
| 3.81908 | 1.00016 | standard |
| 3.80191 | 1.00005 | standard |
| 3.81776 | 1.00005 | standard |
| 3.80244 | 1.001 | standard |
| 3.81724 | 1.001 | standard |
| 3.8029 | 1.00005 | standard |
| 3.81677 | 1.00005 | standard |
| 3.80368 | 1.00016 | standard |
| 3.816 | 1.00016 | standard |
| 3.804 | 1.00014 | standard |
| 3.81568 | 1.00014 | standard |
| 3.80429 | 1.00005 | standard |
| 3.81539 | 1.00005 | standard |
| 3.8048 | 1.00028 | standard |
| 3.81488 | 1.00028 | standard |
| 3.80557 | 1.00001 | standard |
| 3.81411 | 1.00001 | standard |