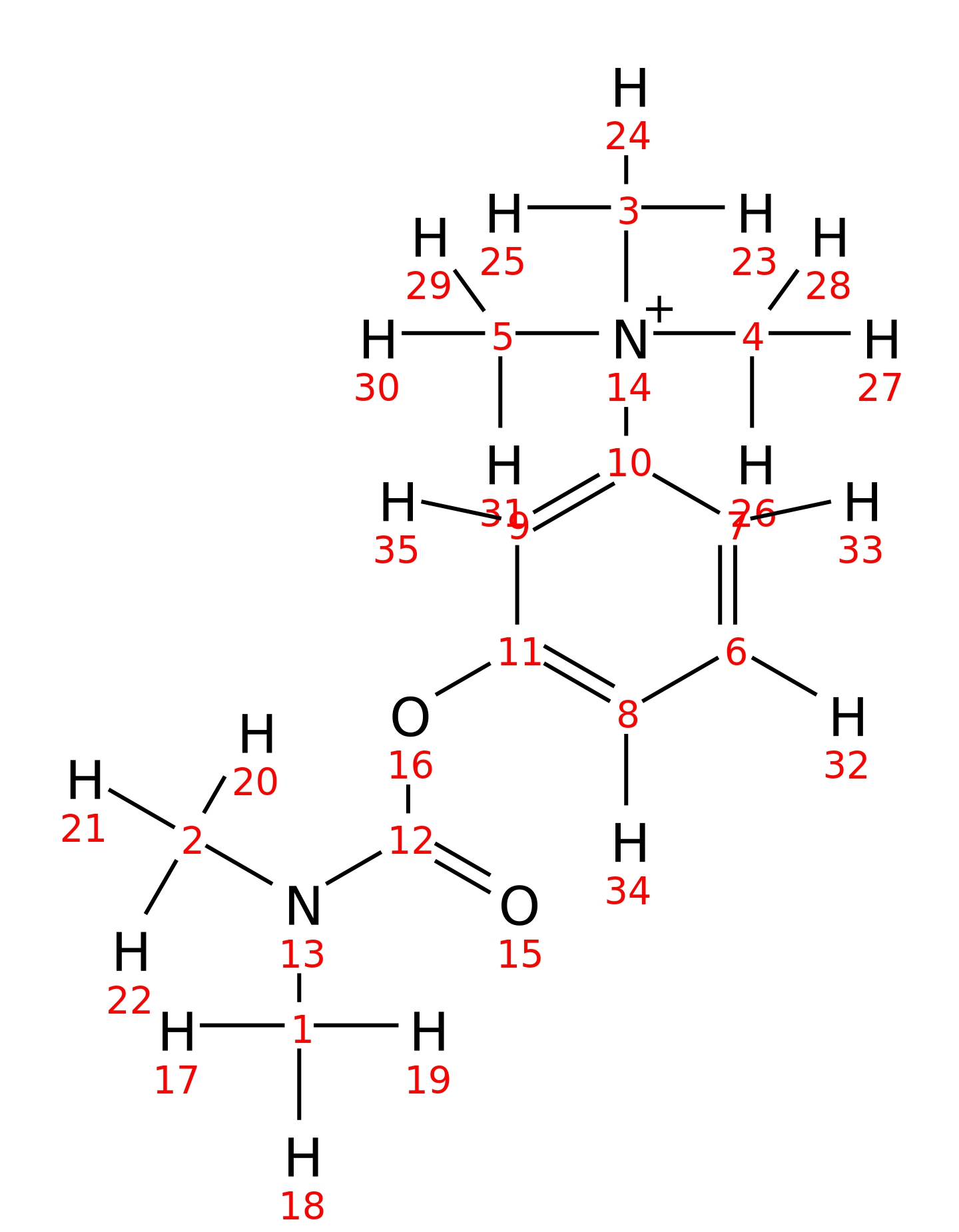
Neostigmine
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01696 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C12H19N2O2/c1-13(2)12(15)16-11-8-6-7-10(9-11)14(3,4)5/h6-9H,1-5H3/q+1 | |
| Note 1 | 33?34 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| neostigmine | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 17 | 18 | 19 | 20 | 21 | 22 | 33 | 32 | 34 | 35 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17 | 3.149 | -12.5 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 3.149 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 3.149 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 3.006 | -12.5 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 3.006 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 3.006 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 33 | 0 | 0 | 0 | 0 | 0 | 0 | 7.389 | 8.205 | 0.856 | 2.401 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.694 | 8.72 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.766 | 2.411 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.702 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.668 | -12.5 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.668 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.668 | 0 | 0 | 0 | 0 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.668 | 0 | 0 | 0 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.668 | -12.5 | 0 | 0 | 0 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.668 | 0 | 0 | 0 |
| 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.668 | -12.5 | -12.5 |
| 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.668 | -12.5 |
| 31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.668 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.00593 | 0.33332 | standard |
| 3.14936 | 0.333495 | standard |
| 3.66751 | 1.0 | standard |
| 7.379 | 0.0268541 | standard |
| 7.3823 | 0.0275114 | standard |
| 7.39524 | 0.0301722 | standard |
| 7.39833 | 0.0299037 | standard |
| 7.67618 | 0.0245914 | standard |
| 7.6935 | 0.0619926 | standard |
| 7.69643 | 0.0501574 | standard |
| 7.70156 | 0.0691387 | standard |
| 7.70634 | 0.0489734 | standard |
| 7.7096 | 0.0459065 | standard |
| 7.75676 | 0.037072 | standard |
| 7.76006 | 0.0339264 | standard |
| 7.77419 | 0.0231621 | standard |
| 7.77732 | 0.021717 | standard |
| 3.00606 | 0.344161 | standard |
| 3.14924 | 0.345018 | standard |
| 3.6675 | 1.0 | standard |
| 7.20563 | 0.00840913 | standard |
| 7.32723 | 0.0208871 | standard |
| 7.40271 | 0.0326881 | standard |
| 7.46163 | 0.0388881 | standard |
| 7.49585 | 0.0481151 | standard |
| 7.68758 | 0.167829 | standard |
| 7.79043 | 0.0727271 | standard |
| 3.00596 | 0.337904 | standard |
| 3.14933 | 0.338917 | standard |
| 3.66751 | 1.0 | standard |
| 7.27655 | 0.0106192 | standard |
| 7.32971 | 0.0209043 | standard |
| 7.36155 | 0.0241521 | standard |
| 7.42305 | 0.0311731 | standard |
| 7.46781 | 0.032823 | standard |
| 7.54654 | 0.0188221 | standard |
| 7.69412 | 0.122734 | standard |
| 7.76792 | 0.0878774 | standard |
| 3.00594 | 0.336243 | standard |
| 3.14935 | 0.336166 | standard |
| 3.66751 | 1.0 | standard |
| 7.30859 | 0.0122851 | standard |
| 7.34345 | 0.02422 | standard |
| 7.36661 | 0.0205821 | standard |
| 7.3807 | 0.0229181 | standard |
| 7.42331 | 0.035767 | standard |
| 7.45023 | 0.0288852 | standard |
| 7.53265 | 0.00686912 | standard |
| 7.57728 | 0.0154631 | standard |
| 7.6847 | 0.0971531 | standard |
| 7.69594 | 0.100243 | standard |
| 7.71967 | 0.104333 | standard |
| 7.75661 | 0.0941097 | standard |
| 7.84618 | 0.00823712 | standard |
| 3.00594 | 0.335622 | standard |
| 3.14935 | 0.335588 | standard |
| 3.66751 | 1.0 | standard |
| 7.25106 | 0.00150513 | standard |
| 7.3187 | 0.0128851 | standard |
| 7.34816 | 0.0251134 | standard |
| 7.36517 | 0.020082 | standard |
| 7.38531 | 0.022966 | standard |
| 7.42126 | 0.0369084 | standard |
| 7.44404 | 0.028061 | standard |
| 7.52214 | 0.00497813 | standard |
| 7.59008 | 0.0157753 | standard |
| 7.68478 | 0.0922192 | standard |
| 7.69697 | 0.093538 | standard |
| 7.72335 | 0.0964411 | standard |
| 7.75319 | 0.096434 | standard |
| 7.81275 | 0.0114211 | standard |
| 7.83785 | 0.00856212 | standard |
| 3.00594 | 0.335221 | standard |
| 3.14935 | 0.33513 | standard |
| 3.66751 | 1.0 | standard |
| 7.26237 | 0.00142713 | standard |
| 7.32661 | 0.013695 | standard |
| 7.35188 | 0.0259 | standard |
| 7.3883 | 0.022707 | standard |
| 7.41936 | 0.0372862 | standard |
| 7.43892 | 0.0275123 | standard |
| 7.47651 | 0.00537113 | standard |
| 7.5143 | 0.00390713 | standard |
| 7.60039 | 0.0162763 | standard |
| 7.68543 | 0.0889031 | standard |
| 7.69796 | 0.0885421 | standard |
| 7.72533 | 0.088317 | standard |
| 7.75082 | 0.0983411 | standard |
| 7.80799 | 0.0113721 | standard |
| 7.83075 | 0.009058 | standard |
| 3.00593 | 0.334051 | standard |
| 3.14936 | 0.33354 | standard |
| 3.66751 | 1.0 | standard |
| 7.37032 | 0.026308 | standard |
| 7.39718 | 0.0239583 | standard |
| 7.40825 | 0.0322602 | standard |
| 7.45995 | 0.00123512 | standard |
| 7.48216 | 0.000861125 | standard |
| 7.6481 | 0.0204036 | standard |
| 7.69076 | 0.0825604 | standard |
| 7.70126 | 0.0755684 | standard |
| 7.71156 | 0.0500767 | standard |
| 7.72955 | 0.0519751 | standard |
| 7.74314 | 0.0466615 | standard |
| 7.75001 | 0.051441 | standard |
| 7.78559 | 0.0139142 | standard |
| 7.79503 | 0.015587 | standard |
| 3.00593 | 0.333602 | standard |
| 3.14936 | 0.333475 | standard |
| 3.66751 | 1.0 | standard |
| 7.37335 | 0.0256348 | standard |
| 7.3774 | 0.0267016 | standard |
| 7.39989 | 0.02967 | standard |
| 7.40364 | 0.0306931 | standard |
| 7.6639 | 0.022692 | standard |
| 7.693 | 0.0815281 | standard |
| 7.70145 | 0.0721121 | standard |
| 7.70885 | 0.0470101 | standard |
| 7.71916 | 0.0442246 | standard |
| 7.75267 | 0.0422548 | standard |
| 7.78196 | 0.018834 | standard |
| 3.00593 | 0.333468 | standard |
| 3.14936 | 0.33343 | standard |
| 3.66751 | 1.0 | standard |
| 7.37669 | 0.0265062 | standard |
| 7.38056 | 0.0274001 | standard |
| 7.3968 | 0.0294121 | standard |
| 7.40059 | 0.0299811 | standard |
| 7.67163 | 0.0240385 | standard |
| 7.69362 | 0.0701562 | standard |
| 7.70153 | 0.0701623 | standard |
| 7.70724 | 0.0468788 | standard |
| 7.71334 | 0.0434947 | standard |
| 7.75503 | 0.0388173 | standard |
| 7.75854 | 0.035178 | standard |
| 7.77681 | 0.0214091 | standard |
| 7.78015 | 0.0200841 | standard |
| 3.00593 | 0.33353 | standard |
| 3.14936 | 0.333284 | standard |
| 3.66751 | 1.0 | standard |
| 7.37896 | 0.027215 | standard |
| 7.3823 | 0.027566 | standard |
| 7.39524 | 0.0301672 | standard |
| 7.39833 | 0.0298988 | standard |
| 7.67618 | 0.0245924 | standard |
| 7.6935 | 0.0619874 | standard |
| 7.69643 | 0.0501512 | standard |
| 7.70156 | 0.0691548 | standard |
| 7.70635 | 0.0491673 | standard |
| 7.70953 | 0.0461072 | standard |
| 7.75676 | 0.037063 | standard |
| 7.76006 | 0.0339173 | standard |
| 7.77418 | 0.0231582 | standard |
| 7.77732 | 0.0217148 | standard |
| 3.00593 | 0.3332 | standard |
| 3.14936 | 0.33357 | standard |
| 3.66751 | 1.0 | standard |
| 7.38052 | 0.0276669 | standard |
| 7.3835 | 0.02805 | standard |
| 7.39407 | 0.0299573 | standard |
| 7.39705 | 0.0300219 | standard |
| 7.67926 | 0.025208 | standard |
| 7.6935 | 0.0584426 | standard |
| 7.69766 | 0.0443264 | standard |
| 7.70158 | 0.068863 | standard |
| 7.70603 | 0.0566451 | standard |
| 7.75806 | 0.0358813 | standard |
| 7.76102 | 0.0333111 | standard |
| 7.77262 | 0.0241853 | standard |
| 7.77537 | 0.0225646 | standard |
| 3.00593 | 0.333791 | standard |
| 3.14936 | 0.332951 | standard |
| 3.66751 | 1.0 | standard |
| 7.38176 | 0.0278258 | standard |
| 7.38423 | 0.0281348 | standard |
| 7.39339 | 0.0297995 | standard |
| 7.39591 | 0.0298002 | standard |
| 7.68139 | 0.0258658 | standard |
| 7.69359 | 0.056556 | standard |
| 7.69827 | 0.0421998 | standard |
| 7.70162 | 0.0694055 | standard |
| 7.70516 | 0.0688712 | standard |
| 7.75911 | 0.0347851 | standard |
| 7.76156 | 0.0326798 | standard |
| 7.77153 | 0.0246502 | standard |
| 7.77405 | 0.023419 | standard |
| 3.00593 | 0.333414 | standard |
| 3.14936 | 0.333318 | standard |
| 3.66751 | 1.0 | standard |
| 7.3823 | 0.0273799 | standard |
| 7.38465 | 0.0284837 | standard |
| 7.393 | 0.0298329 | standard |
| 7.39541 | 0.029872 | standard |
| 7.68223 | 0.0260457 | standard |
| 7.69362 | 0.0555691 | standard |
| 7.69854 | 0.0415591 | standard |
| 7.70166 | 0.0702857 | standard |
| 7.7047 | 0.0705953 | standard |
| 7.75951 | 0.0344091 | standard |
| 7.76188 | 0.032486 | standard |
| 7.77106 | 0.025312 | standard |
| 7.77344 | 0.0239559 | standard |
| 3.00593 | 0.333181 | standard |
| 3.14936 | 0.333544 | standard |
| 3.66751 | 1.0 | standard |
| 7.38268 | 0.0274631 | standard |
| 7.38495 | 0.0285245 | standard |
| 7.39269 | 0.0298402 | standard |
| 7.39514 | 0.0296026 | standard |
| 7.68297 | 0.0262287 | standard |
| 7.69364 | 0.0555623 | standard |
| 7.69874 | 0.0412116 | standard |
| 7.70168 | 0.071757 | standard |
| 7.70421 | 0.0700061 | standard |
| 7.75995 | 0.0334201 | standard |
| 7.7622 | 0.0325606 | standard |
| 7.77067 | 0.0254725 | standard |
| 7.77292 | 0.0237401 | standard |
| 3.00593 | 0.332937 | standard |
| 3.14936 | 0.333775 | standard |
| 3.66751 | 1.0 | standard |
| 7.3834 | 0.0283598 | standard |
| 7.38527 | 0.0287452 | standard |
| 7.39235 | 0.0299331 | standard |
| 7.3944 | 0.0295522 | standard |
| 7.68426 | 0.0264767 | standard |
| 7.69366 | 0.0550218 | standard |
| 7.6991 | 0.0411775 | standard |
| 7.70176 | 0.0777358 | standard |
| 7.76054 | 0.0332782 | standard |
| 7.76257 | 0.0318048 | standard |
| 7.7701 | 0.0266916 | standard |
| 7.77215 | 0.0246138 | standard |
| 3.00593 | 0.333184 | standard |
| 3.14936 | 0.333524 | standard |
| 3.66751 | 1.0 | standard |
| 7.38353 | 0.0280482 | standard |
| 7.38563 | 0.0285186 | standard |
| 7.39215 | 0.0303082 | standard |
| 7.39406 | 0.0291985 | standard |
| 7.68476 | 0.0265736 | standard |
| 7.69367 | 0.0549278 | standard |
| 7.6992 | 0.0411796 | standard |
| 7.70183 | 0.084509 | standard |
| 7.76087 | 0.0326423 | standard |
| 7.76278 | 0.0320806 | standard |
| 7.76993 | 0.0265801 | standard |
| 7.77187 | 0.0251227 | standard |
| 3.00593 | 0.333225 | standard |
| 3.14936 | 0.333479 | standard |
| 3.66751 | 1.0 | standard |
| 7.38388 | 0.0281368 | standard |
| 7.38568 | 0.0285466 | standard |
| 7.39208 | 0.0299924 | standard |
| 7.39388 | 0.029608 | standard |
| 7.6852 | 0.026596 | standard |
| 7.69368 | 0.0549759 | standard |
| 7.69943 | 0.0422591 | standard |
| 7.70178 | 0.0909928 | standard |
| 7.76106 | 0.0329358 | standard |
| 7.76285 | 0.0317065 | standard |
| 7.76975 | 0.026389 | standard |
| 7.77155 | 0.0250225 | standard |
| 3.00593 | 0.333331 | standard |
| 3.14936 | 0.333366 | standard |
| 3.66751 | 1.0 | standard |
| 7.38437 | 0.0282521 | standard |
| 7.38608 | 0.0286918 | standard |
| 7.39178 | 0.0300047 | standard |
| 7.39348 | 0.0295931 | standard |
| 7.68604 | 0.0268506 | standard |
| 7.69375 | 0.0544943 | standard |
| 7.69976 | 0.044655 | standard |
| 7.70153 | 0.0932801 | standard |
| 7.7036 | 0.0431772 | standard |
| 7.76146 | 0.0326139 | standard |
| 7.76315 | 0.0315333 | standard |
| 7.76936 | 0.0267132 | standard |
| 7.77106 | 0.0253721 | standard |
| 3.00593 | 0.333065 | standard |
| 3.14936 | 0.333624 | standard |
| 3.66751 | 1.0 | standard |
| 7.38497 | 0.0290558 | standard |
| 7.38644 | 0.028537 | standard |
| 7.39131 | 0.0294844 | standard |
| 7.39277 | 0.0290629 | standard |
| 7.68723 | 0.0270685 | standard |
| 7.69379 | 0.0531035 | standard |
| 7.70013 | 0.0647461 | standard |
| 7.70157 | 0.0713327 | standard |
| 7.70339 | 0.0400458 | standard |
| 7.76221 | 0.03221 | standard |
| 7.76349 | 0.0314276 | standard |
| 7.7688 | 0.0267329 | standard |
| 7.77026 | 0.0255173 | standard |