5-Methoxypsoralen
Simulation outputs:
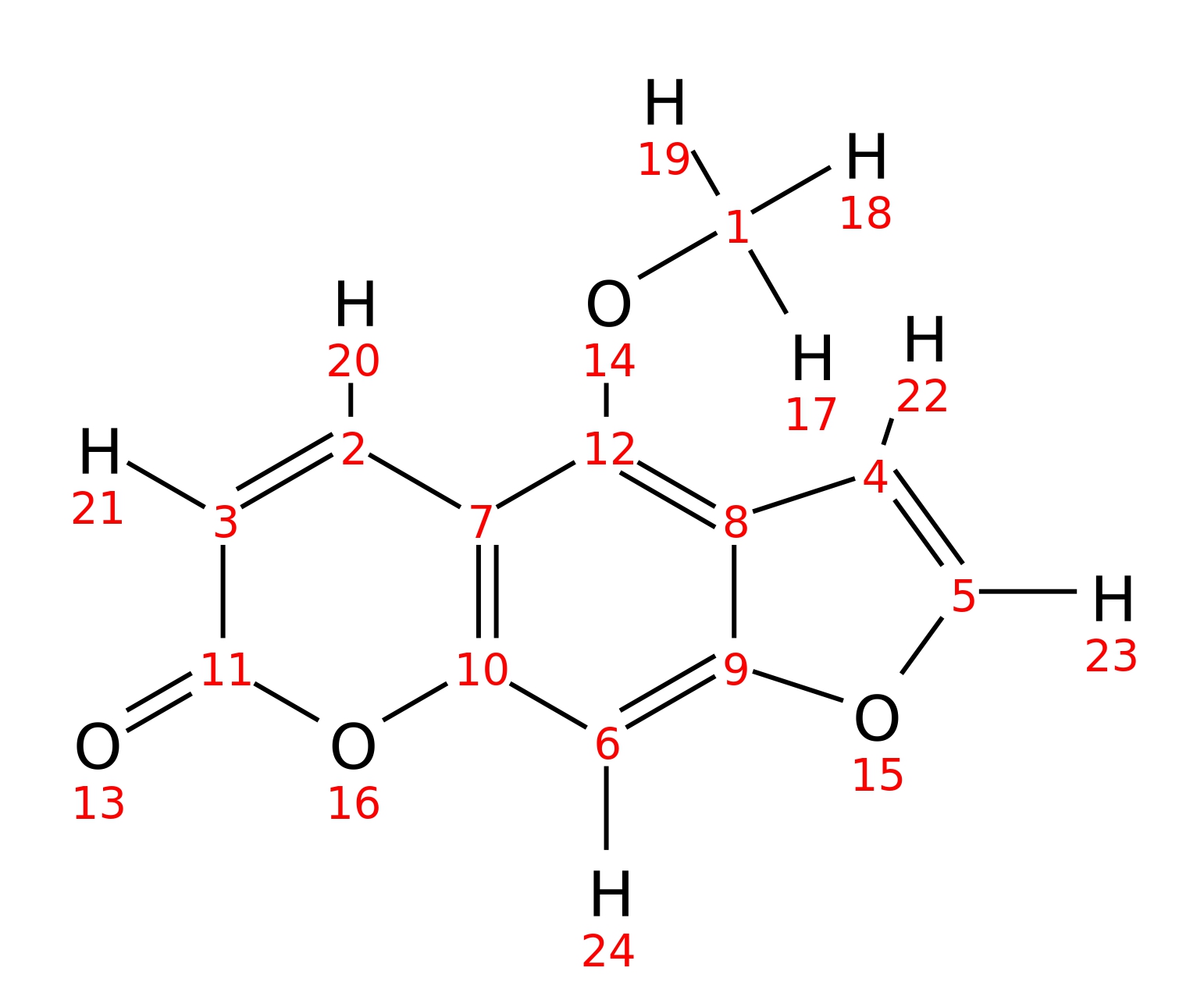
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.02228 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C12H8O4/c1-14-12-7-2-3-11(13)16-10(7)6-9-8(12)4-5-15-9/h2-6H,1H3 | |
| Note 1 | 20?21 ?? 22?23 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 5-Methoxypsoralen | Solute | 100mM |
| CDCl3 | Solvent | 100% |
| TMS | Reference | 1% |
Spin System Matrix
| 17 | 18 | 19 | 20 | 21 | 24 | 22 | 23 | |
|---|---|---|---|---|---|---|---|---|
| 17 | 4.273 | -11.5 | -11.5 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 4.273 | -11.5 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 4.273 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 8.148 | 10.9 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 6.266 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 7.118 | 0.863 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 7.024 | 2.43 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.594 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 4.27334 | 1.0 | standard |
| 6.25468 | 0.167286 | standard |
| 6.2764 | 0.167283 | standard |
| 7.02133 | 0.126661 | standard |
| 7.02239 | 0.132857 | standard |
| 7.02585 | 0.132862 | standard |
| 7.02691 | 0.12667 | standard |
| 7.11803 | 0.238597 | standard |
| 7.11887 | 0.238598 | standard |
| 7.59124 | 0.178243 | standard |
| 7.59605 | 0.178246 | standard |
| 8.13724 | 0.167281 | standard |
| 8.15906 | 0.167278 | standard |
| -0.758733 | 1.125e-06 | standard |
| 4.27334 | 1.0 | standard |
| 6.1196 | 0.143978 | standard |
| 6.39205 | 0.191303 | standard |
| 7.00091 | 0.136624 | standard |
| 7.06112 | 0.182697 | standard |
| 7.11078 | 0.277768 | standard |
| 7.56553 | 0.196093 | standard |
| 7.6248 | 0.1641 | standard |
| 8.02159 | 0.191597 | standard |
| 8.29415 | 0.143907 | standard |
| -0.928243 | 1.125e-06 | standard |
| 4.27334 | 1.0 | standard |
| 6.17038 | 0.151535 | standard |
| 6.35208 | 0.183265 | standard |
| 7.00892 | 0.135096 | standard |
| 7.0483 | 0.152606 | standard |
| 7.11357 | 0.255913 | standard |
| 7.57438 | 0.190054 | standard |
| 7.61425 | 0.168222 | standard |
| 8.06156 | 0.183331 | standard |
| 8.24327 | 0.151543 | standard |
| 4.27334 | 1.0 | standard |
| 6.19497 | 0.155401 | standard |
| 6.33125 | 0.179344 | standard |
| 7.01281 | 0.1344 | standard |
| 7.04212 | 0.14397 | standard |
| 7.11495 | 0.249489 | standard |
| 7.579 | 0.186991 | standard |
| 7.60901 | 0.170559 | standard |
| 8.08239 | 0.179384 | standard |
| 8.21867 | 0.155401 | standard |
| 4.27334 | 1.0 | standard |
| 6.20311 | 0.156698 | standard |
| 6.32421 | 0.177963 | standard |
| 7.01403 | 0.134943 | standard |
| 7.03456 | 0.136464 | standard |
| 7.04016 | 0.140604 | standard |
| 7.11532 | 0.246998 | standard |
| 7.58058 | 0.185274 | standard |
| 7.60729 | 0.171893 | standard |
| 8.08943 | 0.178028 | standard |
| 8.21054 | 0.156668 | standard |
| 4.27334 | 1.0 | standard |
| 6.20955 | 0.157596 | standard |
| 6.31856 | 0.176947 | standard |
| 7.01508 | 0.134489 | standard |
| 7.0334 | 0.136011 | standard |
| 7.03855 | 0.138958 | standard |
| 7.11567 | 0.24536 | standard |
| 7.58185 | 0.184553 | standard |
| 7.60587 | 0.172535 | standard |
| 8.09509 | 0.176967 | standard |
| 8.20409 | 0.157628 | standard |
| 4.27334 | 1.0 | standard |
| 6.23792 | 0.164107 | standard |
| 6.29247 | 0.170515 | standard |
| 7.01966 | 0.134905 | standard |
| 7.02862 | 0.133796 | standard |
| 7.0311 | 0.131818 | standard |
| 7.11709 | 0.238513 | standard |
| 7.11945 | 0.232328 | standard |
| 7.58771 | 0.179219 | standard |
| 7.59969 | 0.177568 | standard |
| 8.12117 | 0.170519 | standard |
| 8.17572 | 0.164108 | standard |
| 4.27334 | 1.0 | standard |
| 6.24724 | 0.167305 | standard |
| 6.28364 | 0.167305 | standard |
| 7.02117 | 0.135232 | standard |
| 7.02703 | 0.133661 | standard |
| 7.02866 | 0.129655 | standard |
| 7.11759 | 0.237678 | standard |
| 7.11916 | 0.234775 | standard |
| 7.58967 | 0.178453 | standard |
| 7.59763 | 0.178452 | standard |
| 8.1301 | 0.16731 | standard |
| 8.1664 | 0.167309 | standard |
| 4.27334 | 1.0 | standard |
| 6.25191 | 0.16729 | standard |
| 6.27918 | 0.16729 | standard |
| 7.02194 | 0.134042 | standard |
| 7.02631 | 0.133586 | standard |
| 7.11793 | 0.240285 | standard |
| 7.11892 | 0.239569 | standard |
| 7.59065 | 0.178341 | standard |
| 7.59664 | 0.178341 | standard |
| 8.13456 | 0.167295 | standard |
| 8.16174 | 0.167295 | standard |
| 4.27334 | 1.0 | standard |
| 6.25468 | 0.167286 | standard |
| 6.2764 | 0.167286 | standard |
| 7.02133 | 0.126638 | standard |
| 7.0224 | 0.132835 | standard |
| 7.02585 | 0.132839 | standard |
| 7.02691 | 0.126645 | standard |
| 7.11803 | 0.238565 | standard |
| 7.11887 | 0.238564 | standard |
| 7.59124 | 0.17824 | standard |
| 7.59605 | 0.17824 | standard |
| 8.13724 | 0.167287 | standard |
| 8.15896 | 0.167286 | standard |
| 4.27334 | 1.0 | standard |
| 6.25647 | 0.167261 | standard |
| 6.27462 | 0.167087 | standard |
| 7.02257 | 0.135192 | standard |
| 7.02567 | 0.135109 | standard |
| 7.118 | 0.232345 | standard |
| 7.11889 | 0.232243 | standard |
| 7.59163 | 0.178137 | standard |
| 7.59566 | 0.177965 | standard |
| 8.13902 | 0.167087 | standard |
| 8.15718 | 0.167261 | standard |
| 4.27334 | 1.0 | standard |
| 6.25776 | 0.16728 | standard |
| 6.27333 | 0.167239 | standard |
| 7.0228 | 0.135304 | standard |
| 7.02544 | 0.135322 | standard |
| 7.11804 | 0.231323 | standard |
| 7.11885 | 0.231342 | standard |
| 7.59193 | 0.178195 | standard |
| 7.59537 | 0.178234 | standard |
| 8.14031 | 0.16728 | standard |
| 8.15589 | 0.167239 | standard |
| 4.27334 | 1.0 | standard |
| 6.25835 | 0.167285 | standard |
| 6.27283 | 0.167284 | standard |
| 7.02223 | 0.126426 | standard |
| 7.02295 | 0.132415 | standard |
| 7.02529 | 0.132416 | standard |
| 7.02601 | 0.126428 | standard |
| 7.11823 | 0.241906 | standard |
| 7.11867 | 0.241906 | standard |
| 7.59203 | 0.178018 | standard |
| 7.59527 | 0.178018 | standard |
| 8.14081 | 0.167285 | standard |
| 8.15529 | 0.167284 | standard |
| 4.27334 | 1.0 | standard |
| 6.25875 | 0.167275 | standard |
| 6.27244 | 0.167275 | standard |
| 7.02305 | 0.134409 | standard |
| 7.02519 | 0.13441 | standard |
| 7.11814 | 0.234834 | standard |
| 7.11875 | 0.234834 | standard |
| 7.59213 | 0.177975 | standard |
| 7.59517 | 0.177975 | standard |
| 8.14131 | 0.167284 | standard |
| 8.15489 | 0.167284 | standard |
| 4.27334 | 1.0 | standard |
| 6.25954 | 0.167281 | standard |
| 6.27164 | 0.16728 | standard |
| 7.02317 | 0.133126 | standard |
| 7.02507 | 0.133126 | standard |
| 7.11831 | 0.243985 | standard |
| 7.11858 | 0.243985 | standard |
| 7.59232 | 0.178415 | standard |
| 7.59497 | 0.178414 | standard |
| 8.142 | 0.16728 | standard |
| 8.1541 | 0.167281 | standard |
| 4.27334 | 1.0 | standard |
| 6.25984 | 0.167276 | standard |
| 6.27135 | 0.167276 | standard |
| 7.02315 | 0.13437 | standard |
| 7.02509 | 0.13437 | standard |
| 7.11823 | 0.237937 | standard |
| 7.11867 | 0.237936 | standard |
| 7.59232 | 0.178061 | standard |
| 7.59488 | 0.178061 | standard |
| 8.1423 | 0.167277 | standard |
| 8.1538 | 0.167277 | standard |
| 4.27334 | 1.0 | standard |
| 6.26014 | 0.167279 | standard |
| 6.27105 | 0.167279 | standard |
| 7.02328 | 0.13362 | standard |
| 7.02496 | 0.13362 | standard |
| 7.11819 | 0.232337 | standard |
| 7.11871 | 0.232337 | standard |
| 7.59242 | 0.177805 | standard |
| 7.59488 | 0.177805 | standard |
| 8.1426 | 0.167279 | standard |
| 8.15351 | 0.167279 | standard |
| 4.27334 | 1.0 | standard |
| 6.26063 | 0.167279 | standard |
| 6.27055 | 0.167278 | standard |
| 7.02327 | 0.132389 | standard |
| 7.02497 | 0.132389 | standard |
| 7.11819 | 0.234633 | standard |
| 7.11861 | 0.234633 | standard |
| 7.59252 | 0.178507 | standard |
| 7.59468 | 0.178506 | standard |
| 8.14309 | 0.167278 | standard |
| 8.15301 | 0.167279 | standard |
| 4.27334 | 1.0 | standard |
| 6.26143 | 0.167287 | standard |
| 6.26976 | 0.167287 | standard |
| 7.02334 | 0.138292 | standard |
| 7.0249 | 0.138292 | standard |
| 7.11824 | 0.231546 | standard |
| 7.11866 | 0.231545 | standard |
| 7.59272 | 0.178061 | standard |
| 7.59458 | 0.178061 | standard |
| 8.14388 | 0.167287 | standard |
| 8.15222 | 0.167287 | standard |