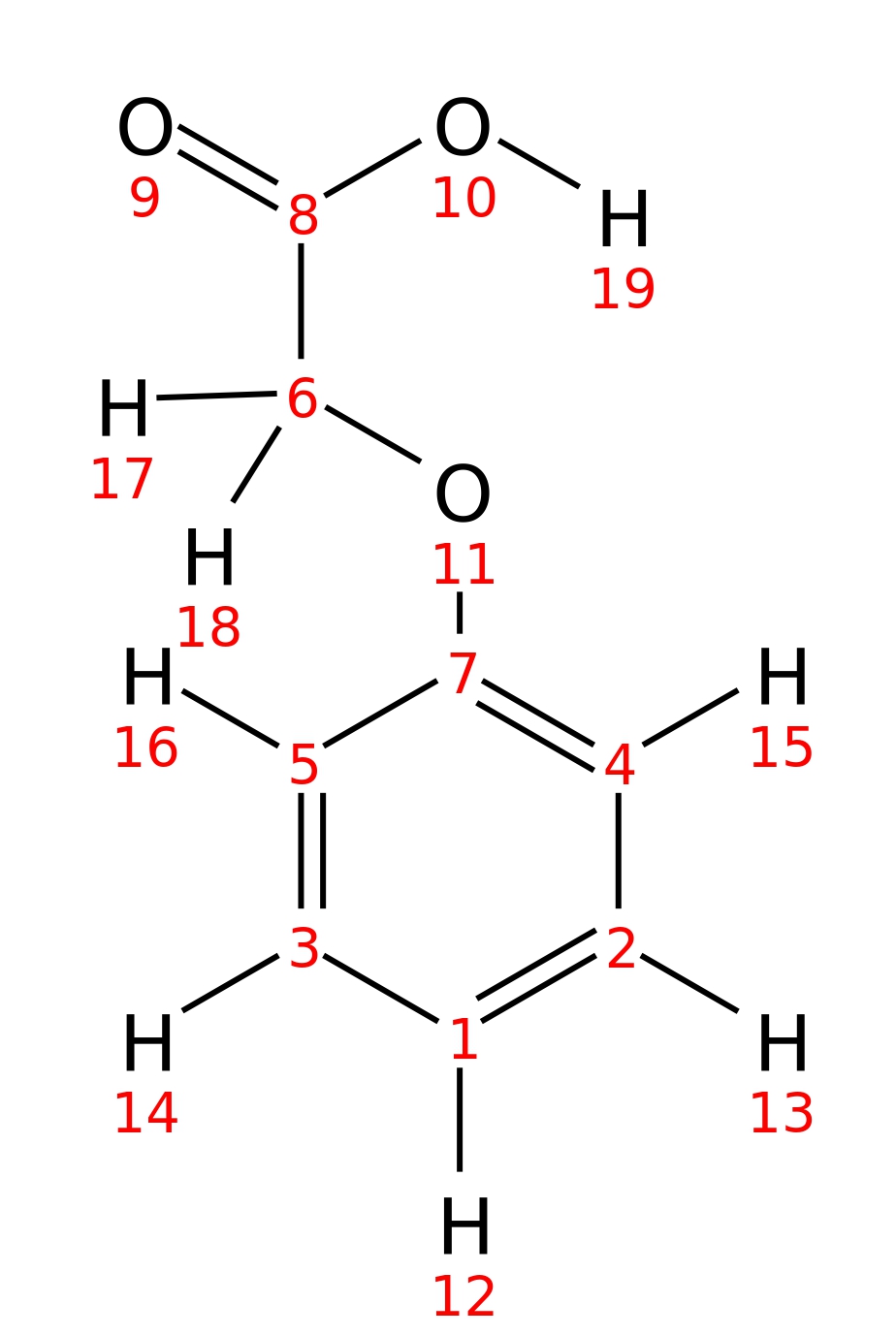
Phenoxyacetic-acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.02470 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H8O3/c9-8(10)6-11-7-4-2-1-3-5-7/h1-5H,6H2,(H,9,10) | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| phenoxyacetic acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 12 | 13 | 15 | 16 | 14 | 17 | 18 | |
|---|---|---|---|---|---|---|---|
| 12 | 7.039 | 7.5 | 0.966 | 0.966 | 7.5 | 0 | 0 |
| 13 | 0 | 7.364 | 7.5 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 6.953 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 6.953 | 7.5 | 0 | 0 |
| 14 | 0 | 0 | 0 | 0 | 7.364 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 4.478 | -12.4 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 4.478 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 4.47826 | 1.0 | standard |
| 6.94489 | 0.330578 | standard |
| 6.9462 | 0.332299 | standard |
| 6.9599 | 0.349866 | standard |
| 6.96119 | 0.352443 | standard |
| 7.02376 | 0.0773076 | standard |
| 7.03874 | 0.167326 | standard |
| 7.05377 | 0.0927892 | standard |
| 7.34896 | 0.271535 | standard |
| 7.36396 | 0.498545 | standard |
| 7.37896 | 0.234151 | standard |
| -0.95542 | 3.125e-06 | standard |
| 4.47826 | 1.0 | standard |
| 6.84286 | 0.156484 | standard |
| 6.88004 | 0.14251 | standard |
| 6.99256 | 0.32792 | standard |
| 7.03364 | 0.556043 | standard |
| 7.13164 | 0.198524 | standard |
| 7.16381 | 0.182241 | standard |
| 7.22652 | 0.453127 | standard |
| 7.31548 | 0.10298 | standard |
| 7.36416 | 0.152308 | standard |
| 7.42536 | 0.141399 | standard |
| 7.55784 | 0.0629732 | standard |
| 7.61239 | 0.0381771 | standard |
| -0.720348 | 1.125e-06 | standard |
| 4.47826 | 1.0 | standard |
| 6.87512 | 0.200787 | standard |
| 6.89283 | 0.218887 | standard |
| 6.99469 | 0.408065 | standard |
| 7.01472 | 0.439087 | standard |
| 7.11797 | 0.134915 | standard |
| 7.13695 | 0.144396 | standard |
| 7.26055 | 0.322808 | standard |
| 7.35481 | 0.13112 | standard |
| 7.38645 | 0.266806 | standard |
| 7.49952 | 0.0889556 | standard |
| -0.880138 | 1.125e-06 | standard |
| 4.47826 | 1.0 | standard |
| 6.90358 | 0.244843 | standard |
| 6.94778 | 0.069658 | standard |
| 7.00014 | 0.42328 | standard |
| 7.02865 | 0.217458 | standard |
| 7.1056 | 0.111898 | standard |
| 7.11801 | 0.13237 | standard |
| 7.2852 | 0.325583 | standard |
| 7.37531 | 0.363306 | standard |
| 7.46565 | 0.115982 | standard |
| -0.961074 | 1.125e-06 | standard |
| 4.47826 | 1.0 | standard |
| 6.90823 | 0.253131 | standard |
| 6.94753 | 0.0823633 | standard |
| 6.9942 | 0.382483 | standard |
| 7.03036 | 0.201152 | standard |
| 7.10063 | 0.104954 | standard |
| 7.11081 | 0.125927 | standard |
| 7.29201 | 0.330399 | standard |
| 7.37276 | 0.388213 | standard |
| 7.45399 | 0.1282 | standard |
| 4.47826 | 1.0 | standard |
| 6.91311 | 0.262682 | standard |
| 6.9594 | 0.09445 | standard |
| 6.98904 | 0.378944 | standard |
| 7.03169 | 0.193042 | standard |
| 7.09625 | 0.0997942 | standard |
| 7.10478 | 0.122277 | standard |
| 7.29787 | 0.32832 | standard |
| 7.37107 | 0.404769 | standard |
| 7.44464 | 0.138705 | standard |
| 4.47826 | 1.0 | standard |
| 6.93232 | 0.299629 | standard |
| 6.93524 | 0.305701 | standard |
| 6.97252 | 0.371166 | standard |
| 6.99962 | 0.075342 | standard |
| 7.03707 | 0.172191 | standard |
| 7.07436 | 0.104754 | standard |
| 7.32825 | 0.297793 | standard |
| 7.36556 | 0.472149 | standard |
| 7.4029 | 0.195645 | standard |
| 4.47826 | 1.0 | standard |
| 6.9393 | 0.315029 | standard |
| 6.94143 | 0.320165 | standard |
| 6.96426 | 0.352544 | standard |
| 6.96639 | 0.36296 | standard |
| 7.01324 | 0.0741356 | standard |
| 7.03823 | 0.170281 | standard |
| 7.06317 | 0.09809 | standard |
| 7.33956 | 0.28407 | standard |
| 7.36449 | 0.488623 | standard |
| 7.38947 | 0.216674 | standard |
| 4.47826 | 1.0 | standard |
| 6.94276 | 0.321641 | standard |
| 6.94445 | 0.324107 | standard |
| 6.96151 | 0.35014 | standard |
| 6.9632 | 0.355885 | standard |
| 7.01986 | 0.0762524 | standard |
| 7.03865 | 0.16857 | standard |
| 7.05732 | 0.0948432 | standard |
| 7.3454 | 0.278149 | standard |
| 7.36411 | 0.494954 | standard |
| 7.38284 | 0.225755 | standard |
| 4.47826 | 1.0 | standard |
| 6.94489 | 0.330535 | standard |
| 6.9462 | 0.332257 | standard |
| 6.9599 | 0.350946 | standard |
| 6.96117 | 0.35294 | standard |
| 7.02376 | 0.0773006 | standard |
| 7.03874 | 0.167308 | standard |
| 7.05377 | 0.0927752 | standard |
| 7.34897 | 0.271554 | standard |
| 7.36396 | 0.498544 | standard |
| 7.37896 | 0.234152 | standard |
| 4.47826 | 1.0 | standard |
| 6.94625 | 0.331404 | standard |
| 6.9474 | 0.331381 | standard |
| 6.95874 | 0.346725 | standard |
| 6.9599 | 0.345515 | standard |
| 7.02641 | 0.0807896 | standard |
| 7.03884 | 0.16926 | standard |
| 7.05139 | 0.089098 | standard |
| 7.35139 | 0.267937 | standard |
| 7.36387 | 0.500179 | standard |
| 7.37636 | 0.239736 | standard |
| 4.47826 | 1.0 | standard |
| 6.94733 | 0.337884 | standard |
| 6.94827 | 0.339501 | standard |
| 6.95802 | 0.345143 | standard |
| 6.95897 | 0.345509 | standard |
| 7.02824 | 0.0815868 | standard |
| 7.03895 | 0.167898 | standard |
| 7.04966 | 0.0882558 | standard |
| 7.35307 | 0.261844 | standard |
| 7.36378 | 0.500995 | standard |
| 7.3745 | 0.244715 | standard |
| 4.47826 | 1.0 | standard |
| 6.94771 | 0.337342 | standard |
| 6.94859 | 0.338308 | standard |
| 6.95769 | 0.344646 | standard |
| 6.95858 | 0.34455 | standard |
| 7.029 | 0.0813455 | standard |
| 7.03894 | 0.167921 | standard |
| 7.04896 | 0.0881598 | standard |
| 7.35377 | 0.261025 | standard |
| 7.36378 | 0.500612 | standard |
| 7.3738 | 0.245051 | standard |
| 4.47826 | 1.0 | standard |
| 6.948 | 0.334767 | standard |
| 6.94889 | 0.335536 | standard |
| 6.95742 | 0.346213 | standard |
| 6.9582 | 0.346051 | standard |
| 7.02963 | 0.0836432 | standard |
| 7.03894 | 0.168045 | standard |
| 7.04837 | 0.0883828 | standard |
| 7.35439 | 0.261399 | standard |
| 7.36377 | 0.500367 | standard |
| 7.37313 | 0.248096 | standard |
| 4.47826 | 1.0 | standard |
| 6.94862 | 0.339467 | standard |
| 6.94934 | 0.340516 | standard |
| 6.95694 | 0.345213 | standard |
| 6.95767 | 0.344919 | standard |
| 7.03072 | 0.0840548 | standard |
| 7.03905 | 0.168386 | standard |
| 7.04738 | 0.0850158 | standard |
| 7.35542 | 0.257681 | standard |
| 7.36374 | 0.501514 | standard |
| 7.37206 | 0.24903 | standard |
| 4.47826 | 1.0 | standard |
| 6.94889 | 0.343836 | standard |
| 6.94954 | 0.344756 | standard |
| 6.95677 | 0.346374 | standard |
| 6.95741 | 0.345459 | standard |
| 7.03111 | 0.0838989 | standard |
| 7.03905 | 0.169155 | standard |
| 7.04687 | 0.0848828 | standard |
| 7.35583 | 0.256366 | standard |
| 7.36371 | 0.501909 | standard |
| 7.37164 | 0.250599 | standard |
| 4.47826 | 1.0 | standard |
| 6.94906 | 0.336388 | standard |
| 6.94976 | 0.337509 | standard |
| 6.95654 | 0.339012 | standard |
| 6.95725 | 0.337896 | standard |
| 7.0315 | 0.0852315 | standard |
| 7.03905 | 0.166352 | standard |
| 7.04653 | 0.0857631 | standard |
| 7.35622 | 0.256122 | standard |
| 7.36371 | 0.50174 | standard |
| 7.3712 | 0.250681 | standard |
| 4.47826 | 1.0 | standard |
| 6.94946 | 0.341331 | standard |
| 6.95004 | 0.343734 | standard |
| 6.95626 | 0.345146 | standard |
| 6.95685 | 0.342738 | standard |
| 7.03221 | 0.0840867 | standard |
| 7.03905 | 0.166984 | standard |
| 7.04589 | 0.0848917 | standard |
| 7.35688 | 0.255263 | standard |
| 7.36371 | 0.501293 | standard |
| 7.37053 | 0.251946 | standard |
| 4.47826 | 1.0 | standard |
| 6.95002 | 0.335774 | standard |
| 6.95056 | 0.338139 | standard |
| 6.95577 | 0.336964 | standard |
| 6.95631 | 0.337586 | standard |
| 7.03329 | 0.0850359 | standard |
| 7.03905 | 0.167466 | standard |
| 7.04479 | 0.0850448 | standard |
| 7.35793 | 0.255428 | standard |
| 7.36368 | 0.503894 | standard |
| 7.36944 | 0.253876 | standard |