N-methyl-L-aspartic-acid
Simulation outputs:
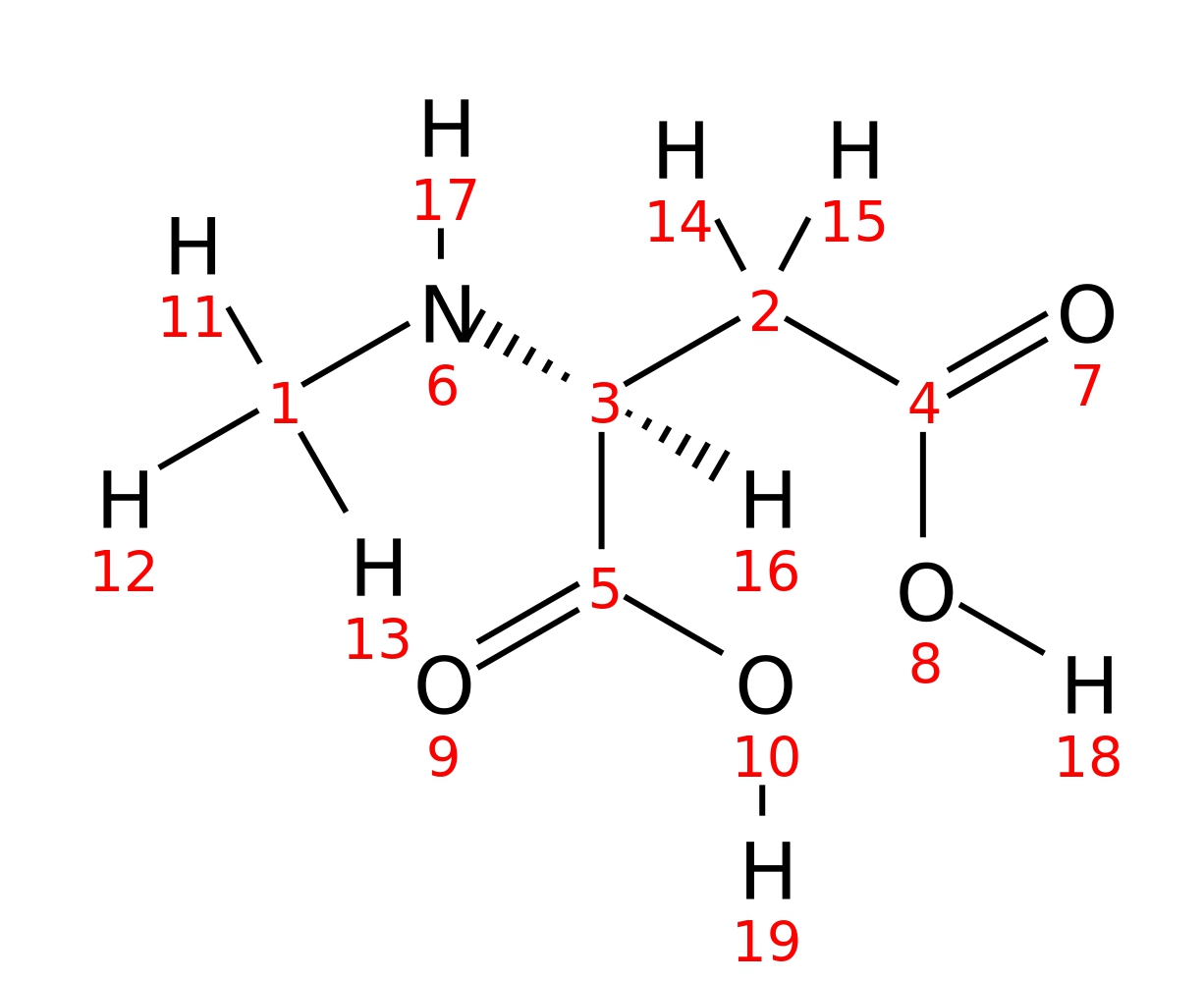
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01214 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C5H9NO4/c1-6-3(5(9)10)2-4(7)8/h3,6H,2H2,1H3,(H,7,8)(H,9,10)/t3-/m0/s1 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N-methyl-L-aspartic acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 11 | 12 | 13 | 16 | 14 | 15 | |
|---|---|---|---|---|---|---|
| 11 | 2.741 | -12.5 | -12.5 | 0 | 0 | 0 |
| 12 | 0 | 2.741 | -12.5 | 0 | 0 | 0 |
| 13 | 0 | 0 | 2.741 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 3.734 | 3.83 | 7.691 |
| 14 | 0 | 0 | 0 | 0 | 2.815 | -17.775 |
| 15 | 0 | 0 | 0 | 0 | 0 | 2.709 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.68031 | 0.058543 | standard |
| 2.69548 | 0.057606 | standard |
| 2.71591 | 0.111924 | standard |
| 2.73111 | 0.12672 | standard |
| 2.7413 | 1.0 | standard |
| 2.79576 | 0.111878 | standard |
| 2.80359 | 0.113954 | standard |
| 2.83127 | 0.0595338 | standard |
| 2.8391 | 0.0577321 | standard |
| 3.72286 | 0.08611 | standard |
| 3.73069 | 0.0882772 | standard |
| 3.73803 | 0.0878422 | standard |
| 3.74596 | 0.0860648 | standard |
| 2.22271 | 0.00479213 | standard |
| 2.66649 | 0.192822 | standard |
| 2.74119 | 1.0 | standard |
| 2.82698 | 0.397918 | standard |
| 3.14613 | 0.00931412 | standard |
| 3.60307 | 0.10739 | standard |
| 3.72552 | 0.092694 | standard |
| 3.76623 | 0.0854992 | standard |
| 3.88872 | 0.0618722 | standard |
| 2.39743 | 0.007453 | standard |
| 2.50886 | 0.00552112 | standard |
| 2.69396 | 0.187536 | standard |
| 2.74095 | 1.00002 | standard |
| 2.80659 | 0.332895 | standard |
| 3.02493 | 0.00998012 | standard |
| 3.64383 | 0.0951668 | standard |
| 3.72468 | 0.087917 | standard |
| 3.7541 | 0.083646 | standard |
| 3.83509 | 0.0655199 | standard |
| 2.48305 | 0.00989212 | standard |
| 2.56788 | 0.00702025 | standard |
| 2.70566 | 0.171596 | standard |
| 2.74155 | 1.00004 | standard |
| 2.79036 | 0.220547 | standard |
| 2.80133 | 0.210736 | standard |
| 2.96532 | 0.0119934 | standard |
| 3.02448 | 0.005754 | standard |
| 3.66535 | 0.0853636 | standard |
| 3.72472 | 0.0813742 | standard |
| 3.74962 | 0.0779222 | standard |
| 3.80902 | 0.0646709 | standard |
| 2.51122 | 0.0122582 | standard |
| 2.58709 | 0.010359 | standard |
| 2.70901 | 0.178812 | standard |
| 2.74156 | 1.00003 | standard |
| 2.78445 | 0.212932 | standard |
| 2.79997 | 0.1979 | standard |
| 2.9458 | 0.0144852 | standard |
| 2.99785 | 0.00760937 | standard |
| 3.67269 | 0.0891078 | standard |
| 3.72488 | 0.0856967 | standard |
| 3.74828 | 0.0824719 | standard |
| 3.80049 | 0.069912 | standard |
| 2.53343 | 0.0144882 | standard |
| 2.60234 | 0.012174 | standard |
| 2.71149 | 0.181568 | standard |
| 2.7414 | 1.0 | standard |
| 2.77984 | 0.210034 | standard |
| 2.79876 | 0.189907 | standard |
| 2.93033 | 0.0167402 | standard |
| 2.97662 | 0.0095765 | standard |
| 3.67863 | 0.0913438 | standard |
| 3.72504 | 0.0883341 | standard |
| 3.74733 | 0.08516 | standard |
| 3.79365 | 0.073405 | standard |
| 2.62943 | 0.0319091 | standard |
| 2.66593 | 0.0300463 | standard |
| 2.71836 | 0.148992 | standard |
| 2.74131 | 1.0 | standard |
| 2.75452 | 0.199765 | standard |
| 2.7756 | 0.146273 | standard |
| 2.79673 | 0.147699 | standard |
| 2.8645 | 0.0339731 | standard |
| 2.88561 | 0.0279216 | standard |
| 3.706 | 0.0884193 | standard |
| 3.72713 | 0.0885784 | standard |
| 3.74237 | 0.086113 | standard |
| 3.7636 | 0.0818614 | standard |
| 2.65868 | 0.041538 | standard |
| 2.68358 | 0.0394871 | standard |
| 2.71791 | 0.120438 | standard |
| 2.74139 | 1.00004 | standard |
| 2.78584 | 0.117926 | standard |
| 2.79932 | 0.120898 | standard |
| 2.84506 | 0.0427043 | standard |
| 2.85863 | 0.0390577 | standard |
| 3.71532 | 0.0803066 | standard |
| 3.72881 | 0.0817766 | standard |
| 3.7402 | 0.0811157 | standard |
| 3.75369 | 0.0780036 | standard |
| 2.67247 | 0.052888 | standard |
| 2.69132 | 0.051401 | standard |
| 2.71691 | 0.118218 | standard |
| 2.73612 | 0.196194 | standard |
| 2.7413 | 1.00001 | standard |
| 2.7918 | 0.117482 | standard |
| 2.8017 | 0.119529 | standard |
| 2.83623 | 0.0537206 | standard |
| 2.84614 | 0.051204 | standard |
| 3.72008 | 0.0854909 | standard |
| 3.7299 | 0.0876942 | standard |
| 3.73892 | 0.0871673 | standard |
| 3.74883 | 0.0854348 | standard |
| 2.68031 | 0.058519 | standard |
| 2.69558 | 0.057618 | standard |
| 2.71591 | 0.112448 | standard |
| 2.73111 | 0.126736 | standard |
| 2.7413 | 1.0 | standard |
| 2.79576 | 0.112407 | standard |
| 2.80359 | 0.113957 | standard |
| 2.83127 | 0.0595138 | standard |
| 2.8391 | 0.0577541 | standard |
| 3.72286 | 0.086134 | standard |
| 3.73069 | 0.0882552 | standard |
| 3.73803 | 0.0882992 | standard |
| 3.74586 | 0.086135 | standard |
| 2.68546 | 0.062414 | standard |
| 2.69816 | 0.061816 | standard |
| 2.71502 | 0.107821 | standard |
| 2.72772 | 0.113239 | standard |
| 2.7413 | 1.0 | standard |
| 2.79854 | 0.108428 | standard |
| 2.80498 | 0.109612 | standard |
| 2.8281 | 0.0634919 | standard |
| 2.83463 | 0.0621411 | standard |
| 3.72474 | 0.0861999 | standard |
| 3.73128 | 0.0883722 | standard |
| 3.73743 | 0.0883712 | standard |
| 3.74397 | 0.0862219 | standard |
| 2.68903 | 0.065263 | standard |
| 2.69994 | 0.06488 | standard |
| 2.71443 | 0.104483 | standard |
| 2.72534 | 0.107276 | standard |
| 2.7413 | 1.0 | standard |
| 2.80052 | 0.105369 | standard |
| 2.80607 | 0.106298 | standard |
| 2.82591 | 0.066453 | standard |
| 2.83146 | 0.0654182 | standard |
| 3.72613 | 0.0862961 | standard |
| 3.73168 | 0.0883751 | standard |
| 3.73703 | 0.0883741 | standard |
| 3.74259 | 0.086296 | standard |
| 2.69042 | 0.066434 | standard |
| 2.70054 | 0.066126 | standard |
| 2.71413 | 0.103147 | standard |
| 2.72424 | 0.105234 | standard |
| 2.7413 | 1.0 | standard |
| 2.80141 | 0.104157 | standard |
| 2.80657 | 0.10499 | standard |
| 2.82512 | 0.067668 | standard |
| 2.83027 | 0.0667453 | standard |
| 3.72663 | 0.0862429 | standard |
| 3.73188 | 0.0883711 | standard |
| 3.73684 | 0.0884391 | standard |
| 3.74199 | 0.0863351 | standard |
| 2.69161 | 0.067462 | standard |
| 2.70113 | 0.067223 | standard |
| 2.71383 | 0.101967 | standard |
| 2.72335 | 0.103584 | standard |
| 2.7413 | 1.0 | standard |
| 2.80211 | 0.103036 | standard |
| 2.80696 | 0.103788 | standard |
| 2.82433 | 0.068714 | standard |
| 2.82918 | 0.0678813 | standard |
| 3.72712 | 0.086304 | standard |
| 3.73198 | 0.0883251 | standard |
| 3.73674 | 0.088402 | standard |
| 3.7415 | 0.0864053 | standard |
| 2.6936 | 0.069194 | standard |
| 2.70213 | 0.068754 | standard |
| 2.71333 | 0.099997 | standard |
| 2.72186 | 0.101024 | standard |
| 2.7413 | 1.0 | standard |
| 2.8034 | 0.101127 | standard |
| 2.80776 | 0.101307 | standard |
| 2.82314 | 0.070474 | standard |
| 2.82749 | 0.0697842 | standard |
| 3.72791 | 0.0862719 | standard |
| 3.73228 | 0.0883641 | standard |
| 3.73644 | 0.0884401 | standard |
| 3.7407 | 0.0859992 | standard |
| 2.69449 | 0.069944 | standard |
| 2.70252 | 0.069855 | standard |
| 2.71314 | 0.099166 | standard |
| 2.72127 | 0.1 | standard |
| 2.7413 | 1.0 | standard |
| 2.80399 | 0.100437 | standard |
| 2.80805 | 0.101011 | standard |
| 2.82264 | 0.0712971 | standard |
| 2.8267 | 0.0706654 | standard |
| 3.72831 | 0.0863531 | standard |
| 3.73237 | 0.0884101 | standard |
| 3.73634 | 0.0884091 | standard |
| 3.7404 | 0.0863531 | standard |
| 2.69518 | 0.070614 | standard |
| 2.70282 | 0.070557 | standard |
| 2.71294 | 0.098418 | standard |
| 2.72067 | 0.0991 | standard |
| 2.7413 | 1.0 | standard |
| 2.80449 | 0.0997188 | standard |
| 2.80835 | 0.10025 | standard |
| 2.82224 | 0.0719831 | standard |
| 2.82611 | 0.0714014 | standard |
| 3.72861 | 0.0863491 | standard |
| 3.73247 | 0.0884061 | standard |
| 3.73624 | 0.0884051 | standard |
| 3.74011 | 0.0863491 | standard |
| 2.69647 | 0.071784 | standard |
| 2.70342 | 0.071783 | standard |
| 2.71264 | 0.097149 | standard |
| 2.71958 | 0.097533 | standard |
| 2.7413 | 1.0 | standard |
| 2.80528 | 0.0984077 | standard |
| 2.80885 | 0.0988685 | standard |
| 2.82145 | 0.0731341 | standard |
| 2.82502 | 0.0725763 | standard |
| 3.7291 | 0.0863423 | standard |
| 3.73257 | 0.08839 | standard |
| 3.73604 | 0.0882851 | standard |
| 3.73961 | 0.08627 | standard |
| 2.69846 | 0.07361 | standard |
| 2.70431 | 0.073679 | standard |
| 2.71214 | 0.09518 | standard |
| 2.718 | 0.095382 | standard |
| 2.7413 | 1.0 | standard |
| 2.80667 | 0.0965658 | standard |
| 2.80964 | 0.0969177 | standard |
| 2.82036 | 0.0750672 | standard |
| 2.82333 | 0.0746754 | standard |
| 3.7299 | 0.0863441 | standard |
| 3.73287 | 0.088305 | standard |
| 3.73585 | 0.088435 | standard |
| 3.73872 | 0.0865076 | standard |