3-Methylphenylacetate
Simulation outputs:
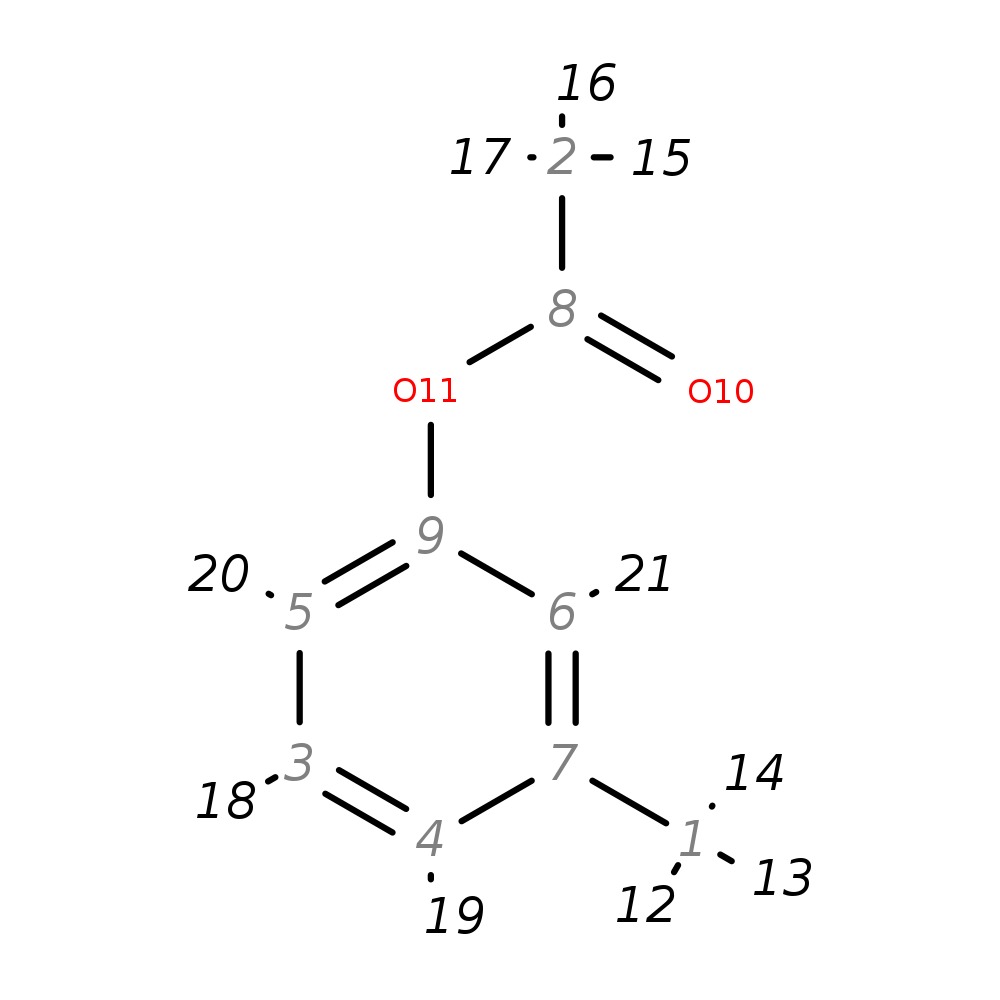
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01622 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H10O2/c1-7-4-3-5-9(6-7)11-8(2)10/h3-6H,1-2H3 | |
| Note 1 | Looks like NMR contains contamination. Fitted by splitting matrix. | |
| Note 2 | Oscar Li |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-methylphenylacetate | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 2.324 | 0 | -12.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 2.324 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 2.324 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 2.347 | -12.4 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 2.347 | -12.4 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 2.347 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 7.349 | 7.847 | 7.95 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.179 | 1.33 | 1.33 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.942 | 1.33 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.989 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.9232 | standard |
| 6.93381 | 0.0766661 | standard |
| 6.94964 | 0.0828702 | standard |
| 6.98931 | 0.153564 | standard |
| 7.17091 | 0.0725194 | standard |
| 7.18658 | 0.0860445 | standard |
| 7.3337 | 0.0619707 | standard |
| 7.34945 | 0.10577 | standard |
| 7.36525 | 0.0484532 | standard |
| 2.33002 | 1.0 | standard |
| 7.01222 | 0.165174 | standard |
| 7.23132 | 0.152727 | standard |
| 7.40361 | 0.0425301 | standard |
| 7.59603 | 0.0107321 | standard |
| 2.32562 | 1.0 | standard |
| 2.34448 | 0.93622 | standard |
| 6.87604 | 0.0440761 | standard |
| 6.89905 | 0.0452941 | standard |
| 6.99694 | 0.192565 | standard |
| 7.10258 | 0.0407331 | standard |
| 7.22676 | 0.115045 | standard |
| 7.25351 | 0.130119 | standard |
| 7.37712 | 0.0589493 | standard |
| 7.50627 | 0.0159641 | standard |
| 2.32456 | 1.00001 | standard |
| 2.34605 | 0.927869 | standard |
| 6.88992 | 0.0537383 | standard |
| 6.99079 | 0.21459 | standard |
| 7.12251 | 0.0414931 | standard |
| 7.214 | 0.101935 | standard |
| 7.27362 | 0.093406 | standard |
| 7.36595 | 0.070601 | standard |
| 7.46128 | 0.021181 | standard |
| 2.32434 | 1.0 | standard |
| 2.34633 | 0.926272 | standard |
| 6.89561 | 0.057309 | standard |
| 6.98895 | 0.218153 | standard |
| 7.12817 | 0.0439551 | standard |
| 7.21117 | 0.10048 | standard |
| 7.28008 | 0.088226 | standard |
| 7.36302 | 0.0754092 | standard |
| 7.44755 | 0.0239401 | standard |
| 2.32421 | 1.0 | standard |
| 2.34649 | 0.925312 | standard |
| 6.90029 | 0.0601 | standard |
| 6.98731 | 0.216416 | standard |
| 7.13303 | 0.0454641 | standard |
| 7.20887 | 0.099409 | standard |
| 7.28525 | 0.0853192 | standard |
| 7.3608 | 0.080238 | standard |
| 7.437 | 0.026422 | standard |
| 2.32395 | 1.00003 | standard |
| 2.34682 | 0.923406 | standard |
| 6.92142 | 0.071477 | standard |
| 6.96084 | 0.0931819 | standard |
| 6.98917 | 0.157312 | standard |
| 7.15746 | 0.0612051 | standard |
| 7.19633 | 0.0933392 | standard |
| 7.31309 | 0.0719453 | standard |
| 7.35213 | 0.100124 | standard |
| 7.39113 | 0.0394789 | standard |
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.923247 | standard |
| 6.92837 | 0.0746053 | standard |
| 6.95467 | 0.0859313 | standard |
| 6.98927 | 0.15462 | standard |
| 7.16505 | 0.0674617 | standard |
| 7.19109 | 0.0898353 | standard |
| 7.32419 | 0.0665306 | standard |
| 7.35039 | 0.103825 | standard |
| 7.37658 | 0.044305 | standard |
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.923213 | standard |
| 6.93176 | 0.0757972 | standard |
| 6.95159 | 0.0838707 | standard |
| 6.98927 | 0.1544 | standard |
| 7.16879 | 0.0708681 | standard |
| 7.18835 | 0.087319 | standard |
| 7.33008 | 0.0637051 | standard |
| 7.34975 | 0.105069 | standard |
| 7.36941 | 0.0468794 | standard |
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.9232 | standard |
| 6.93388 | 0.0767271 | standard |
| 6.94964 | 0.0828662 | standard |
| 6.98931 | 0.153548 | standard |
| 7.17091 | 0.0725123 | standard |
| 7.18658 | 0.0860325 | standard |
| 7.3337 | 0.0619677 | standard |
| 7.34945 | 0.105776 | standard |
| 7.3652 | 0.0484523 | standard |
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.923193 | standard |
| 6.9352 | 0.0771551 | standard |
| 6.94842 | 0.0822734 | standard |
| 6.98932 | 0.154716 | standard |
| 7.17234 | 0.0736896 | standard |
| 7.18531 | 0.0850698 | standard |
| 7.33613 | 0.0608251 | standard |
| 7.34931 | 0.105921 | standard |
| 7.36243 | 0.0495269 | standard |
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.923188 | standard |
| 6.93618 | 0.0771769 | standard |
| 6.94751 | 0.0821826 | standard |
| 6.98933 | 0.154333 | standard |
| 7.17332 | 0.0742642 | standard |
| 7.18448 | 0.0839448 | standard |
| 7.33791 | 0.0600042 | standard |
| 7.34921 | 0.106038 | standard |
| 7.36049 | 0.050313 | standard |
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.923187 | standard |
| 6.93653 | 0.0776226 | standard |
| 6.94711 | 0.0816043 | standard |
| 6.98934 | 0.153561 | standard |
| 7.17372 | 0.0755483 | standard |
| 7.18414 | 0.0839788 | standard |
| 7.3387 | 0.0597224 | standard |
| 7.34916 | 0.106355 | standard |
| 7.3597 | 0.050624 | standard |
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.923185 | standard |
| 6.93692 | 0.0775706 | standard |
| 6.94681 | 0.0812801 | standard |
| 6.98934 | 0.153183 | standard |
| 7.17406 | 0.0750168 | standard |
| 7.18384 | 0.0835129 | standard |
| 7.3393 | 0.0594044 | standard |
| 7.34916 | 0.106395 | standard |
| 7.35901 | 0.050916 | standard |
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.923183 | standard |
| 6.93746 | 0.0787311 | standard |
| 6.94626 | 0.0820557 | standard |
| 6.98934 | 0.153571 | standard |
| 7.17465 | 0.0756885 | standard |
| 7.18335 | 0.0832658 | standard |
| 7.34039 | 0.0589705 | standard |
| 7.34908 | 0.106262 | standard |
| 7.35782 | 0.051392 | standard |
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.923182 | standard |
| 6.93771 | 0.0784195 | standard |
| 6.94601 | 0.081543 | standard |
| 6.98934 | 0.154349 | standard |
| 7.17485 | 0.0762604 | standard |
| 7.1831 | 0.0824411 | standard |
| 7.34078 | 0.0587415 | standard |
| 7.34906 | 0.10647 | standard |
| 7.35742 | 0.0515258 | standard |
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.923181 | standard |
| 6.93789 | 0.0795026 | standard |
| 6.94579 | 0.0796164 | standard |
| 6.98936 | 0.153567 | standard |
| 7.17503 | 0.076071 | standard |
| 7.1829 | 0.0828188 | standard |
| 7.34118 | 0.0575105 | standard |
| 7.34906 | 0.106566 | standard |
| 7.35693 | 0.0527339 | standard |
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.92318 | standard |
| 6.9383 | 0.0801231 | standard |
| 6.94553 | 0.0791372 | standard |
| 6.98935 | 0.154757 | standard |
| 7.17544 | 0.0769561 | standard |
| 7.18256 | 0.0832118 | standard |
| 7.34187 | 0.0572765 | standard |
| 7.34906 | 0.106553 | standard |
| 7.35624 | 0.0529328 | standard |
| 2.32393 | 1.0 | standard |
| 2.34684 | 0.923178 | standard |
| 6.9389 | 0.0793746 | standard |
| 6.94492 | 0.0805676 | standard |
| 6.98937 | 0.153035 | standard |
| 7.17604 | 0.0778632 | standard |
| 7.18208 | 0.0818674 | standard |
| 7.34296 | 0.0569345 | standard |
| 7.34906 | 0.106554 | standard |
| 7.35505 | 0.0533421 | standard |