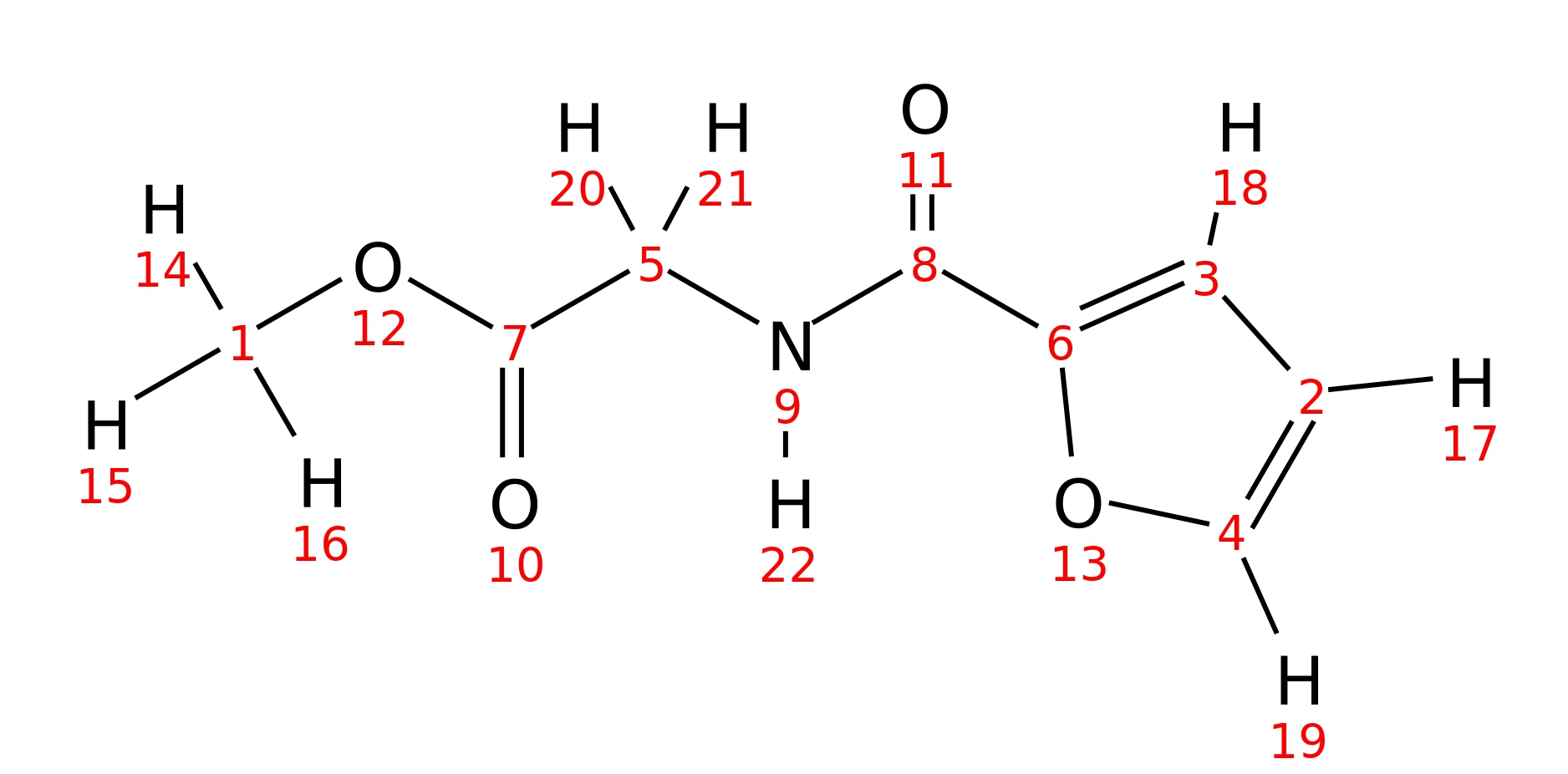
N-2-furoyl-glycine-methyl-ester
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.00916 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H9NO4/c1-12-7(10)5-9-8(11)6-3-2-4-13-6/h2-4H,5H2,1H3,(H,9,11) | |
| Note 1 | 18?19 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N-(2-furoyl)glycine methyl ester | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 14 | 15 | 16 | 20 | 21 | 18 | 17 | 19 | |
|---|---|---|---|---|---|---|---|---|
| 14 | 3.78 | -11.5 | -11.5 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 3.78 | -11.5 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 3.78 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 4.177 | -12.4 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 4.177 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 7.203 | 3.402 | 0.84 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 6.636 | 1.845 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.691 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666672 | standard |
| 6.63084 | 0.0951277 | standard |
| 6.63449 | 0.104938 | standard |
| 6.63751 | 0.104639 | standard |
| 6.64127 | 0.0946638 | standard |
| 7.19865 | 0.1186 | standard |
| 7.19978 | 0.120391 | standard |
| 7.20543 | 0.120391 | standard |
| 7.20657 | 0.118601 | standard |
| 7.69036 | 0.140256 | standard |
| 7.69257 | 0.142321 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.66749 | standard |
| 6.56753 | 0.0802801 | standard |
| 6.61329 | 0.0948036 | standard |
| 6.65181 | 0.113307 | standard |
| 6.69737 | 0.111705 | standard |
| 7.15667 | 0.132959 | standard |
| 7.17096 | 0.138617 | standard |
| 7.24048 | 0.103535 | standard |
| 7.25522 | 0.103248 | standard |
| 7.66293 | 0.132828 | standard |
| 7.6794 | 0.142934 | standard |
| 7.70439 | 0.140028 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666979 | standard |
| 6.59111 | 0.0860461 | standard |
| 6.62169 | 0.0975763 | standard |
| 6.64734 | 0.111287 | standard |
| 6.67777 | 0.105022 | standard |
| 7.17107 | 0.12891 | standard |
| 7.18063 | 0.132031 | standard |
| 7.22718 | 0.109579 | standard |
| 7.23683 | 0.107812 | standard |
| 7.67295 | 0.130173 | standard |
| 7.6832 | 0.142257 | standard |
| 7.70014 | 0.141151 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666735 | standard |
| 6.60261 | 0.0883394 | standard |
| 6.62556 | 0.0988717 | standard |
| 6.64478 | 0.110156 | standard |
| 6.66763 | 0.101968 | standard |
| 7.1787 | 0.126889 | standard |
| 7.18581 | 0.12891 | standard |
| 7.22072 | 0.112084 | standard |
| 7.22805 | 0.110242 | standard |
| 7.68517 | 0.141689 | standard |
| 7.698 | 0.141407 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666769 | standard |
| 6.60645 | 0.0895503 | standard |
| 6.62682 | 0.0996051 | standard |
| 6.64389 | 0.10953 | standard |
| 6.66419 | 0.100755 | standard |
| 7.18128 | 0.125706 | standard |
| 7.18755 | 0.127561 | standard |
| 7.21872 | 0.113867 | standard |
| 7.225 | 0.111893 | standard |
| 7.68592 | 0.141064 | standard |
| 7.69731 | 0.141555 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666679 | standard |
| 6.60939 | 0.089883 | standard |
| 6.62779 | 0.100097 | standard |
| 6.6431 | 0.10904 | standard |
| 6.66143 | 0.100121 | standard |
| 7.18341 | 0.125567 | standard |
| 7.18894 | 0.127258 | standard |
| 7.21713 | 0.114684 | standard |
| 7.22266 | 0.112861 | standard |
| 7.6864 | 0.141328 | standard |
| 7.69669 | 0.141198 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666687 | standard |
| 6.62287 | 0.0937957 | standard |
| 6.63206 | 0.103427 | standard |
| 6.63976 | 0.105554 | standard |
| 6.64894 | 0.0962076 | standard |
| 7.19287 | 0.120754 | standard |
| 7.19563 | 0.1225 | standard |
| 7.20964 | 0.118463 | standard |
| 7.21259 | 0.116558 | standard |
| 7.68883 | 0.141498 | standard |
| 7.69407 | 0.141507 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666638 | standard |
| 6.6273 | 0.0950026 | standard |
| 6.63342 | 0.104564 | standard |
| 6.63849 | 0.104741 | standard |
| 6.64461 | 0.0952219 | standard |
| 7.19609 | 0.119303 | standard |
| 7.19792 | 0.121171 | standard |
| 7.20725 | 0.119311 | standard |
| 7.20928 | 0.117266 | standard |
| 7.68766 | 0.126832 | standard |
| 7.6898 | 0.141097 | standard |
| 7.69309 | 0.141112 | standard |
| 7.69523 | 0.126839 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666321 | standard |
| 6.62946 | 0.094808 | standard |
| 6.63411 | 0.104497 | standard |
| 6.6379 | 0.104483 | standard |
| 6.64255 | 0.0948028 | standard |
| 7.19774 | 0.119179 | standard |
| 7.19912 | 0.120986 | standard |
| 7.2061 | 0.1211 | standard |
| 7.20747 | 0.1193 | standard |
| 7.69017 | 0.142149 | standard |
| 7.69272 | 0.142243 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666671 | standard |
| 6.63084 | 0.0951207 | standard |
| 6.63449 | 0.104926 | standard |
| 6.63751 | 0.104627 | standard |
| 6.64127 | 0.0946568 | standard |
| 7.19865 | 0.118582 | standard |
| 7.19978 | 0.120373 | standard |
| 7.20543 | 0.120373 | standard |
| 7.20657 | 0.118582 | standard |
| 7.69036 | 0.14023 | standard |
| 7.69257 | 0.142295 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.66667 | standard |
| 6.63173 | 0.095019 | standard |
| 6.63479 | 0.104768 | standard |
| 6.63732 | 0.104768 | standard |
| 6.64038 | 0.095019 | standard |
| 7.1993 | 0.119182 | standard |
| 7.20021 | 0.120947 | standard |
| 7.20486 | 0.117852 | standard |
| 7.20596 | 0.115803 | standard |
| 7.69057 | 0.141497 | standard |
| 7.69232 | 0.141497 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666698 | standard |
| 6.63231 | 0.094758 | standard |
| 6.63499 | 0.104787 | standard |
| 6.63712 | 0.104787 | standard |
| 6.63979 | 0.094758 | standard |
| 7.19976 | 0.119256 | standard |
| 7.20054 | 0.120964 | standard |
| 7.20458 | 0.120964 | standard |
| 7.20535 | 0.119256 | standard |
| 7.69067 | 0.140404 | standard |
| 7.69223 | 0.140404 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666669 | standard |
| 6.63251 | 0.0948392 | standard |
| 6.63498 | 0.104428 | standard |
| 6.63703 | 0.104428 | standard |
| 6.6395 | 0.0948392 | standard |
| 7.19991 | 0.115847 | standard |
| 7.20078 | 0.118041 | standard |
| 7.20448 | 0.121908 | standard |
| 7.20515 | 0.120265 | standard |
| 7.68986 | 0.125042 | standard |
| 7.69082 | 0.140901 | standard |
| 7.69211 | 0.143519 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666672 | standard |
| 6.63281 | 0.0951527 | standard |
| 6.63508 | 0.104413 | standard |
| 6.63703 | 0.104412 | standard |
| 6.6393 | 0.0951428 | standard |
| 7.20011 | 0.116864 | standard |
| 7.20088 | 0.118929 | standard |
| 7.20433 | 0.118926 | standard |
| 7.20511 | 0.116854 | standard |
| 7.68996 | 0.125711 | standard |
| 7.69079 | 0.139286 | standard |
| 7.69214 | 0.142441 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666668 | standard |
| 6.6331 | 0.0947726 | standard |
| 6.63518 | 0.104926 | standard |
| 6.63683 | 0.104926 | standard |
| 6.63891 | 0.0947726 | standard |
| 7.20044 | 0.121091 | standard |
| 7.20096 | 0.122526 | standard |
| 7.20412 | 0.117809 | standard |
| 7.20484 | 0.115809 | standard |
| 7.69013 | 0.124873 | standard |
| 7.69091 | 0.140385 | standard |
| 7.69202 | 0.143646 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666668 | standard |
| 6.6333 | 0.0946935 | standard |
| 6.63528 | 0.104987 | standard |
| 6.63683 | 0.104987 | standard |
| 6.63881 | 0.0946926 | standard |
| 7.20049 | 0.11827 | standard |
| 7.20109 | 0.120276 | standard |
| 7.20403 | 0.120276 | standard |
| 7.20463 | 0.11827 | standard |
| 7.69088 | 0.13978 | standard |
| 7.69201 | 0.13978 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666668 | standard |
| 6.6334 | 0.094588 | standard |
| 6.63527 | 0.10402 | standard |
| 6.63684 | 0.104653 | standard |
| 6.63861 | 0.0955424 | standard |
| 7.20056 | 0.115656 | standard |
| 7.20121 | 0.117777 | standard |
| 7.20404 | 0.123146 | standard |
| 7.20448 | 0.121779 | standard |
| 7.69087 | 0.143563 | standard |
| 7.69189 | 0.139996 | standard |
| 7.69258 | 0.124704 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666667 | standard |
| 6.63369 | 0.0949561 | standard |
| 6.63537 | 0.10496 | standard |
| 6.63673 | 0.10496 | standard |
| 6.63841 | 0.0949561 | standard |
| 7.20075 | 0.116889 | standard |
| 7.20131 | 0.118726 | standard |
| 7.20396 | 0.124989 | standard |
| 7.20426 | 0.123959 | standard |
| 7.69044 | 0.12606 | standard |
| 7.691 | 0.140625 | standard |
| 7.69189 | 0.140625 | standard |
| 7.69246 | 0.12606 | standard |
| 3.77959 | 1.0 | standard |
| 4.17722 | 0.666627 | standard |
| 6.63399 | 0.0941994 | standard |
| 6.63547 | 0.104184 | standard |
| 6.63664 | 0.104935 | standard |
| 6.63802 | 0.0953899 | standard |
| 7.20103 | 0.115263 | standard |
| 7.20153 | 0.117277 | standard |
| 7.20373 | 0.124701 | standard |
| 7.20398 | 0.123121 | standard |
| 7.69096 | 0.143305 | standard |
| 7.69179 | 0.138225 | standard |
| 7.69235 | 0.123842 | standard |