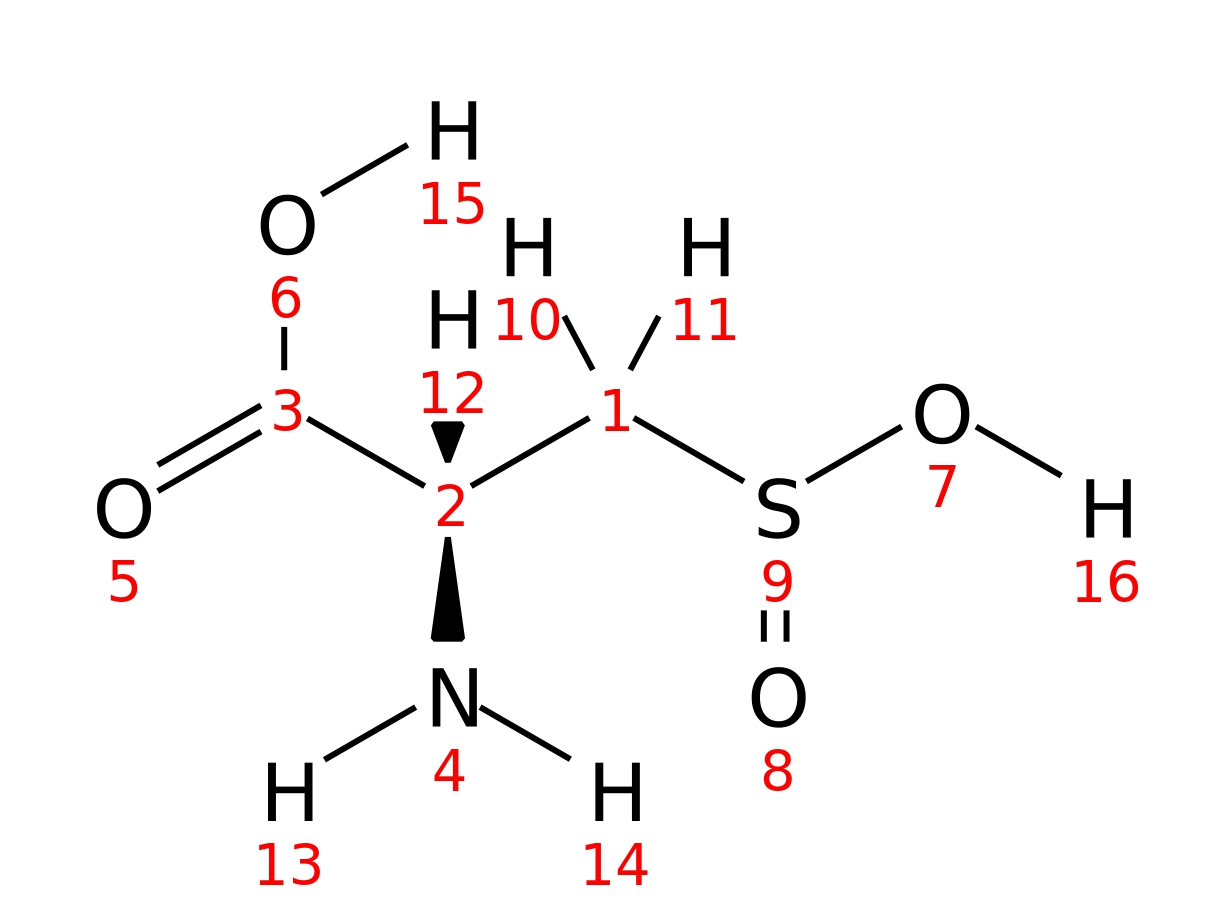
L-cysteinesulfinic-acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.02542 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C3H7NO4S/c4-2(3(5)6)1-9(7)8/h2H,1,4H2,(H,5,6)(H,7,8)/t2-/m0/s1 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| L-cysteinesulfinic acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 10 | 11 | 12 | |
|---|---|---|---|
| 10 | 2.693 | -14.2 | 10.04 |
| 11 | 0 | 2.812 | 2.35 |
| 12 | 0 | 0 | 4.139 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.66711 | 0.606126 | standard |
| 2.68695 | 0.597166 | standard |
| 2.69548 | 0.951 | standard |
| 2.71542 | 0.964811 | standard |
| 2.79735 | 0.978877 | standard |
| 2.8023 | 1.00005 | standard |
| 2.82582 | 0.6297 | standard |
| 2.83066 | 0.608221 | standard |
| 4.12665 | 0.805932 | standard |
| 4.13149 | 0.807941 | standard |
| 4.14648 | 0.80793 | standard |
| 4.15133 | 0.805933 | standard |
| 2.2808 | 0.026779 | standard |
| 2.63376 | 0.376268 | standard |
| 2.70617 | 0.4034 | standard |
| 2.82373 | 1.0 | standard |
| 3.05895 | 0.0462716 | standard |
| 3.76248 | 0.0240324 | standard |
| 3.99682 | 0.280458 | standard |
| 4.11338 | 0.230846 | standard |
| 4.18778 | 0.207542 | standard |
| 4.30436 | 0.184249 | standard |
| 4.53861 | 0.00887725 | standard |
| 2.42848 | 0.047974 | standard |
| 2.56289 | 0.0166141 | standard |
| 2.66455 | 0.435003 | standard |
| 2.7327 | 0.463216 | standard |
| 2.80151 | 1.0 | standard |
| 2.96871 | 0.0645985 | standard |
| 3.0401 | 0.00836512 | standard |
| 3.87117 | 0.0183282 | standard |
| 4.04115 | 0.305438 | standard |
| 4.11276 | 0.268455 | standard |
| 4.17533 | 0.246021 | standard |
| 4.24704 | 0.230096 | standard |
| 4.41691 | 0.00929925 | standard |
| 2.50058 | 0.101492 | standard |
| 2.6059 | 0.0467522 | standard |
| 2.67783 | 0.668441 | standard |
| 2.7474 | 0.721072 | standard |
| 2.78347 | 1.0 | standard |
| 2.79629 | 0.98583 | standard |
| 2.92459 | 0.118554 | standard |
| 2.97392 | 0.0308725 | standard |
| 3.92293 | 0.0179261 | standard |
| 4.06456 | 0.45623 | standard |
| 4.11384 | 0.420997 | standard |
| 4.16979 | 0.391915 | standard |
| 4.21907 | 0.374944 | standard |
| 4.36067 | 0.0118675 | standard |
| 2.52409 | 0.123393 | standard |
| 2.61946 | 0.0639059 | standard |
| 2.6817 | 0.715583 | standard |
| 2.7527 | 0.788832 | standard |
| 2.7771 | 1.0 | standard |
| 2.79461 | 0.96967 | standard |
| 2.91021 | 0.141217 | standard |
| 2.95232 | 0.0464054 | standard |
| 3.9394 | 0.0167252 | standard |
| 4.0725 | 0.489897 | standard |
| 4.11463 | 0.457797 | standard |
| 4.16781 | 0.427649 | standard |
| 4.20994 | 0.411656 | standard |
| 4.34303 | 0.0113513 | standard |
| 2.54284 | 0.136596 | standard |
| 2.63006 | 0.0790945 | standard |
| 2.68467 | 0.717196 | standard |
| 2.75718 | 0.847034 | standard |
| 2.77178 | 1.00002 | standard |
| 2.79341 | 0.930263 | standard |
| 2.89881 | 0.155874 | standard |
| 2.93527 | 0.0613277 | standard |
| 3.95199 | 0.0145642 | standard |
| 4.07894 | 0.493761 | standard |
| 4.11542 | 0.46622 | standard |
| 4.16612 | 0.436955 | standard |
| 4.2026 | 0.430724 | standard |
| 2.62337 | 0.321185 | standard |
| 2.67099 | 0.284648 | standard |
| 2.69429 | 0.872384 | standard |
| 2.7419 | 1.0 | standard |
| 2.7796 | 0.920362 | standard |
| 2.79386 | 0.995208 | standard |
| 2.85052 | 0.344589 | standard |
| 2.86487 | 0.268086 | standard |
| 4.1084 | 0.655073 | standard |
| 4.12276 | 0.646266 | standard |
| 4.15601 | 0.604223 | standard |
| 4.17037 | 0.6115 | standard |
| 2.64827 | 0.445622 | standard |
| 2.6809 | 0.434393 | standard |
| 2.69558 | 0.908467 | standard |
| 2.72821 | 0.987937 | standard |
| 2.78893 | 0.952253 | standard |
| 2.79753 | 1.00004 | standard |
| 2.83623 | 0.472889 | standard |
| 2.84494 | 0.42476 | standard |
| 4.11852 | 0.730981 | standard |
| 4.12713 | 0.727995 | standard |
| 4.15105 | 0.695237 | standard |
| 4.15976 | 0.697649 | standard |
| 2.66017 | 0.543689 | standard |
| 2.68487 | 0.525208 | standard |
| 2.69568 | 0.941691 | standard |
| 2.72038 | 0.965875 | standard |
| 2.79408 | 0.970127 | standard |
| 2.80031 | 1.00005 | standard |
| 2.82959 | 0.563477 | standard |
| 2.83582 | 0.533102 | standard |
| 4.12357 | 0.76747 | standard |
| 4.1298 | 0.769305 | standard |
| 4.14827 | 0.769286 | standard |
| 4.1545 | 0.767472 | standard |
| 2.66711 | 0.606225 | standard |
| 2.68695 | 0.597268 | standard |
| 2.69548 | 0.951027 | standard |
| 2.71542 | 0.964831 | standard |
| 2.79735 | 0.978888 | standard |
| 2.8023 | 1.00005 | standard |
| 2.82582 | 0.629789 | standard |
| 2.83066 | 0.60832 | standard |
| 4.12665 | 0.80598 | standard |
| 4.13149 | 0.807988 | standard |
| 4.14648 | 0.807978 | standard |
| 4.15133 | 0.805981 | standard |
| 2.67158 | 0.652535 | standard |
| 2.68824 | 0.649525 | standard |
| 2.69528 | 0.95617 | standard |
| 2.71194 | 0.963425 | standard |
| 2.79963 | 0.984346 | standard |
| 2.80369 | 1.00006 | standard |
| 2.82334 | 0.677884 | standard |
| 2.82739 | 0.66188 | standard |
| 4.12863 | 0.832106 | standard |
| 4.13268 | 0.834222 | standard |
| 4.14529 | 0.835755 | standard |
| 4.14925 | 0.833694 | standard |
| 2.67485 | 0.688725 | standard |
| 2.68903 | 0.689375 | standard |
| 2.69508 | 0.95991 | standard |
| 2.70937 | 0.962945 | standard |
| 2.80132 | 0.987891 | standard |
| 2.80478 | 1.00006 | standard |
| 2.82165 | 0.717309 | standard |
| 2.82501 | 0.704939 | standard |
| 4.13012 | 0.852896 | standard |
| 4.13358 | 0.85509 | standard |
| 4.1444 | 0.85696 | standard |
| 4.14776 | 0.854829 | standard |
| 2.67604 | 0.702811 | standard |
| 2.68943 | 0.705311 | standard |
| 2.69498 | 0.960289 | standard |
| 2.70828 | 0.961415 | standard |
| 2.80201 | 0.989268 | standard |
| 2.80517 | 1.00007 | standard |
| 2.82096 | 0.731589 | standard |
| 2.82412 | 0.720556 | standard |
| 4.13071 | 0.861554 | standard |
| 4.13387 | 0.863726 | standard |
| 4.144 | 0.863723 | standard |
| 4.14717 | 0.861555 | standard |
| 2.67713 | 0.716445 | standard |
| 2.68963 | 0.719853 | standard |
| 2.69488 | 0.961392 | standard |
| 2.70738 | 0.961327 | standard |
| 2.80261 | 0.99037 | standard |
| 2.80557 | 1.00007 | standard |
| 2.82036 | 0.745794 | standard |
| 2.82333 | 0.735876 | standard |
| 4.13121 | 0.869215 | standard |
| 4.13417 | 0.871392 | standard |
| 4.14371 | 0.871389 | standard |
| 4.14667 | 0.869216 | standard |
| 2.67902 | 0.738374 | standard |
| 2.69013 | 0.743667 | standard |
| 2.69479 | 0.961596 | standard |
| 2.70589 | 0.959326 | standard |
| 2.8037 | 0.992158 | standard |
| 2.80627 | 1.00008 | standard |
| 2.81947 | 0.77018 | standard |
| 2.82204 | 0.762066 | standard |
| 4.1321 | 0.88236 | standard |
| 4.13467 | 0.884506 | standard |
| 4.14321 | 0.884504 | standard |
| 4.14578 | 0.88236 | standard |
| 2.67971 | 0.749314 | standard |
| 2.69032 | 0.755671 | standard |
| 2.69469 | 0.963806 | standard |
| 2.7052 | 0.960432 | standard |
| 2.80409 | 0.992809 | standard |
| 2.80656 | 1.00007 | standard |
| 2.81907 | 0.780717 | standard |
| 2.82154 | 0.773269 | standard |
| 4.1324 | 0.88796 | standard |
| 4.13487 | 0.890154 | standard |
| 4.14291 | 0.887531 | standard |
| 4.14548 | 0.885259 | standard |
| 2.6804 | 0.759175 | standard |
| 2.69042 | 0.766022 | standard |
| 2.69459 | 0.965185 | standard |
| 2.70461 | 0.961137 | standard |
| 2.80449 | 0.993371 | standard |
| 2.80686 | 1.00007 | standard |
| 2.81867 | 0.790353 | standard |
| 2.82105 | 0.783474 | standard |
| 4.13279 | 0.896195 | standard |
| 4.13507 | 0.898366 | standard |
| 4.14271 | 0.895333 | standard |
| 4.14508 | 0.893077 | standard |
| 2.68159 | 0.775915 | standard |
| 2.69072 | 0.783924 | standard |
| 2.69449 | 0.967086 | standard |
| 2.70361 | 0.961674 | standard |
| 2.80518 | 0.99432 | standard |
| 2.80736 | 1.00007 | standard |
| 2.81808 | 0.807299 | standard |
| 2.82025 | 0.801384 | standard |
| 4.13329 | 0.90201 | standard |
| 4.13546 | 0.904283 | standard |
| 4.14241 | 0.904282 | standard |
| 4.14459 | 0.90201 | standard |
| 2.68338 | 0.799615 | standard |
| 2.69112 | 0.809159 | standard |
| 2.69429 | 0.966365 | standard |
| 2.70203 | 0.959067 | standard |
| 2.80628 | 0.995726 | standard |
| 2.80805 | 1.00008 | standard |
| 2.81719 | 0.834163 | standard |
| 2.81896 | 0.829668 | standard |
| 4.13418 | 0.916195 | standard |
| 4.13596 | 0.918392 | standard |
| 4.14192 | 0.918392 | standard |
| 4.1437 | 0.916195 | standard |