4-Hydroxy-3-methoxymandelic-acid
Simulation outputs:
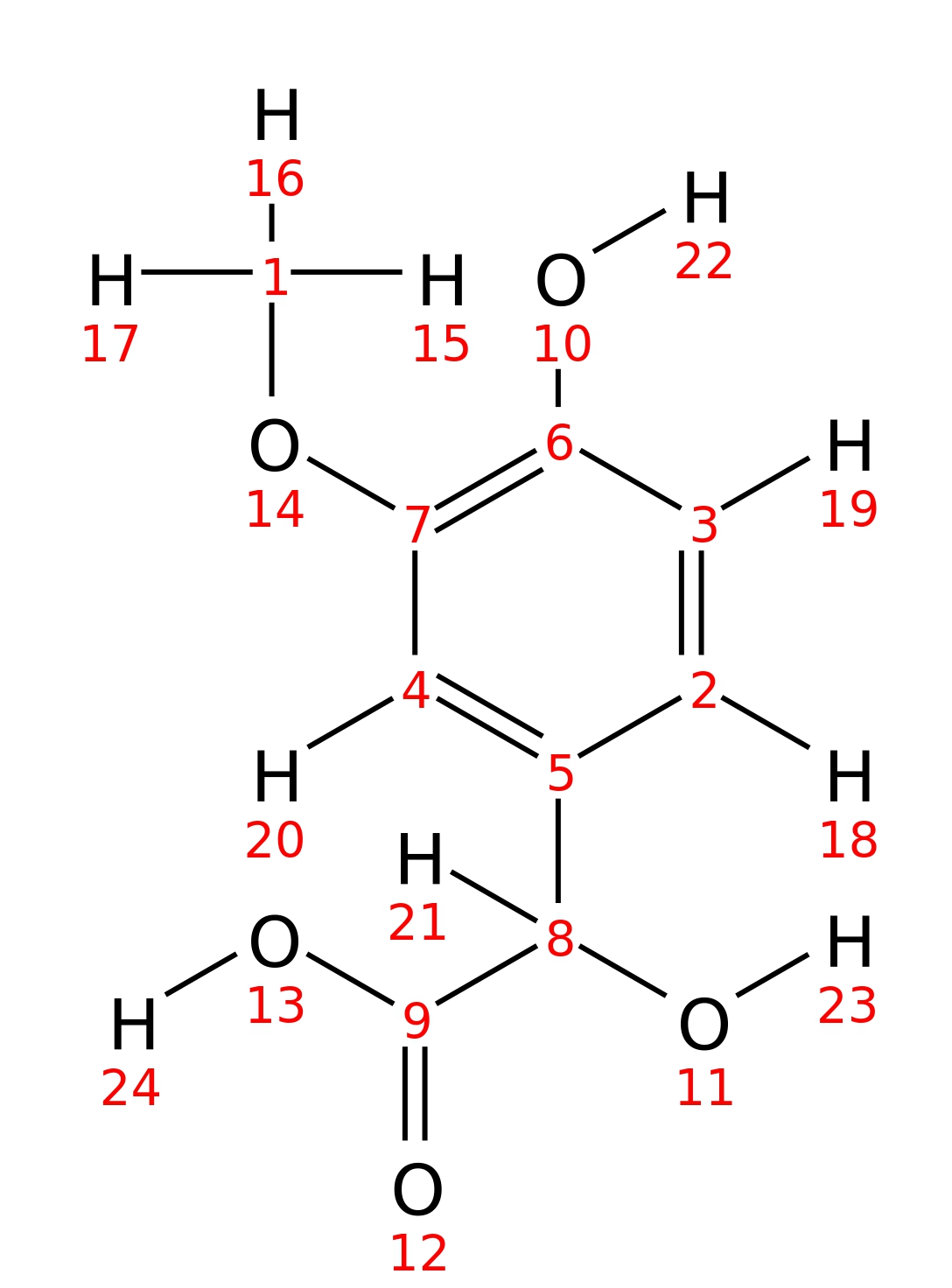
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01571 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H10O5/c1-14-7-4-5(2-3-6(7)10)8(11)9(12)13/h2-4,8,10-11H,1H3,(H,12,13)/t8-/m1/s1 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4-hydroxy-3-methoxymandelic acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 15 | 16 | 17 | 19 | 18 | 20 | 21 | |
|---|---|---|---|---|---|---|---|
| 15 | 3.867 | -11.5 | -11.5 | 0 | 0 | 0 | 0 |
| 16 | 0 | 3.867 | -11.5 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 3.867 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 6.897 | 8.32 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 6.913 | 2.016 | 1.0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 7.048 | 1.0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 4.907 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.86697 | 1.0 | standard |
| 4.90711 | 0.254848 | standard |
| 6.88515 | 0.0501618 | standard |
| 6.9018 | 0.313414 | standard |
| 6.90659 | 0.21233 | standard |
| 6.92543 | 0.0349228 | standard |
| 7.04776 | 0.207313 | standard |
| 3.86697 | 1.0 | standard |
| 4.90732 | 0.259722 | standard |
| 6.90882 | 0.55644 | standard |
| 7.03979 | 0.255938 | standard |
| 3.86697 | 1.0 | standard |
| 4.90727 | 0.259136 | standard |
| 6.90857 | 0.512582 | standard |
| 7.0435 | 0.227929 | standard |
| 3.86697 | 1.0 | standard |
| 4.90696 | 0.258053 | standard |
| 6.90522 | 0.506059 | standard |
| 7.04559 | 0.22155 | standard |
| 3.86697 | 1.0 | standard |
| 4.9069 | 0.257831 | standard |
| 6.90488 | 0.508808 | standard |
| 7.04602 | 0.2221 | standard |
| 3.86697 | 1.0 | standard |
| 4.90696 | 0.258047 | standard |
| 6.90469 | 0.510973 | standard |
| 7.04632 | 0.222736 | standard |
| 3.86697 | 1.0 | standard |
| 4.90702 | 0.257103 | standard |
| 6.8638 | 0.0190391 | standard |
| 6.9039 | 0.496565 | standard |
| 6.95105 | 0.0181441 | standard |
| 7.04725 | 0.219705 | standard |
| 3.86697 | 1.0 | standard |
| 4.90705 | 0.256809 | standard |
| 6.8756 | 0.0283052 | standard |
| 6.90335 | 0.447647 | standard |
| 6.93632 | 0.0237473 | standard |
| 7.04758 | 0.214612 | standard |
| 3.86697 | 1.0 | standard |
| 4.90707 | 0.255471 | standard |
| 6.88158 | 0.0389538 | standard |
| 6.90263 | 0.366758 | standard |
| 6.9293 | 0.0295491 | standard |
| 7.04768 | 0.209886 | standard |
| 3.86697 | 1.0 | standard |
| 4.90711 | 0.25482 | standard |
| 6.88515 | 0.0501746 | standard |
| 6.9018 | 0.31337 | standard |
| 6.90659 | 0.212263 | standard |
| 6.92543 | 0.0349249 | standard |
| 7.04776 | 0.20728 | standard |
| 3.86697 | 1.0 | standard |
| 4.90711 | 0.252314 | standard |
| 6.88744 | 0.060901 | standard |
| 6.9013 | 0.290138 | standard |
| 6.90767 | 0.185386 | standard |
| 6.92281 | 0.0399373 | standard |
| 7.04774 | 0.20217 | standard |
| 3.86697 | 1.0 | standard |
| 4.90711 | 0.252983 | standard |
| 6.88906 | 0.0701412 | standard |
| 6.90091 | 0.275396 | standard |
| 6.90836 | 0.16944 | standard |
| 6.92114 | 0.0441702 | standard |
| 7.04774 | 0.19703 | standard |
| 3.86697 | 1.0 | standard |
| 4.90711 | 0.253413 | standard |
| 6.8897 | 0.0744922 | standard |
| 6.90076 | 0.268649 | standard |
| 6.90859 | 0.165651 | standard |
| 6.92041 | 0.0471886 | standard |
| 7.04779 | 0.195241 | standard |
| 3.86697 | 1.0 | standard |
| 4.90715 | 0.25185 | standard |
| 6.89024 | 0.0788114 | standard |
| 6.90064 | 0.264344 | standard |
| 6.90885 | 0.160264 | standard |
| 6.91972 | 0.0482932 | standard |
| 7.04772 | 0.194813 | standard |
| 3.86697 | 1.0 | standard |
| 4.90714 | 0.254776 | standard |
| 6.8911 | 0.0861493 | standard |
| 6.90034 | 0.254423 | standard |
| 6.90904 | 0.15403 | standard |
| 6.91896 | 0.052203 | standard |
| 7.04784 | 0.196058 | standard |
| 3.86697 | 1.0 | standard |
| 4.90711 | 0.252635 | standard |
| 6.89147 | 0.089474 | standard |
| 6.90021 | 0.25141 | standard |
| 6.90916 | 0.147975 | standard |
| 6.91848 | 0.0539087 | standard |
| 7.04791 | 0.193171 | standard |
| 3.86697 | 1.0 | standard |
| 4.90711 | 0.252507 | standard |
| 6.89179 | 0.0923175 | standard |
| 6.90009 | 0.248048 | standard |
| 6.90945 | 0.146372 | standard |
| 6.91799 | 0.0559209 | standard |
| 7.04788 | 0.193237 | standard |
| 3.86697 | 1.0 | standard |
| 4.9071 | 0.251166 | standard |
| 6.89233 | 0.0982032 | standard |
| 6.89989 | 0.241399 | standard |
| 6.90942 | 0.141143 | standard |
| 6.91701 | 0.0582392 | standard |
| 7.04773 | 0.19277 | standard |
| 3.86697 | 1.0 | standard |
| 4.90715 | 0.250191 | standard |
| 6.89315 | 0.10755 | standard |
| 6.89954 | 0.231609 | standard |
| 6.90982 | 0.132706 | standard |
| 6.91036 | 0.132116 | standard |
| 6.91651 | 0.0633232 | standard |
| 7.04772 | 0.188941 | standard |