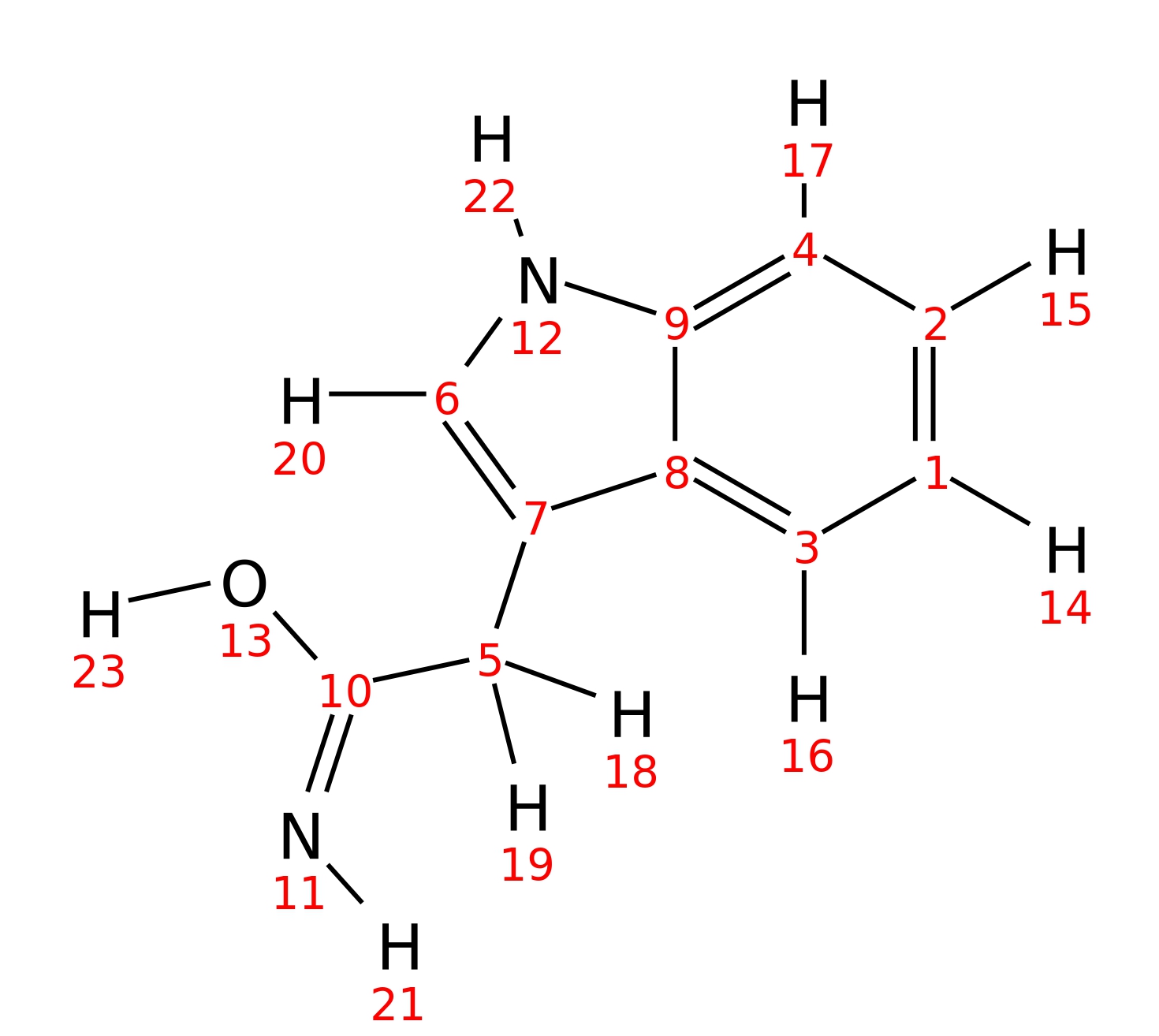
Indole-3-acetamide
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01600 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H10N2O/c11-10(13)5-7-6-12-9-4-2-1-3-8(7)9/h1-4,6,12H,5H2,(H2,11,13) | |
| Note 1 | 14.16?15,17 | |
| Note 2 | Ethanol 5.3204,3.5540,1.1095 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| indole-3-acetamide | Solute | Saturated1 |
| ethanol | Solvent | 100% |
| TMS | Reference | 0.5% |
Spin System Matrix
| 14 | 15 | 16 | 17 | 20 | 18 | 19 | |
|---|---|---|---|---|---|---|---|
| 14 | 6.981 | 7.245 | 0 | 7.51 | 0 | 0 | 0 |
| 15 | 0 | 7.064 | 7.84 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 7.341 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 7.532 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 7.154 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 3.634 | -12.4 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 3.634 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.63378 | 1.0 | standard |
| 6.96572 | 0.109676 | standard |
| 6.98037 | 0.248162 | standard |
| 6.9951 | 0.156253 | standard |
| 7.04982 | 0.147659 | standard |
| 7.065 | 0.238829 | standard |
| 7.07985 | 0.118424 | standard |
| 7.1545 | 0.500505 | standard |
| 7.33379 | 0.267878 | standard |
| 7.34932 | 0.240164 | standard |
| 7.52472 | 0.258582 | standard |
| 7.53959 | 0.251947 | standard |
| -0.938757 | 3.125e-06 | standard |
| 3.63378 | 1.0 | standard |
| 6.77824 | 0.0366771 | standard |
| 6.92751 | 0.193759 | standard |
| 7.02278 | 0.215462 | standard |
| 7.1038 | 0.527518 | standard |
| 7.15391 | 0.686313 | standard |
| 7.26756 | 0.327523 | standard |
| 7.45416 | 0.267329 | standard |
| 7.54118 | 0.136829 | standard |
| 7.63903 | 0.123988 | standard |
| -0.809914 | 1.125e-06 | standard |
| 3.63378 | 1.0 | standard |
| 6.84196 | 0.039769 | standard |
| 6.95589 | 0.230745 | standard |
| 7.01127 | 0.234838 | standard |
| 7.07226 | 0.371374 | standard |
| 7.15462 | 0.567365 | standard |
| 7.29016 | 0.242361 | standard |
| 7.35593 | 0.130396 | standard |
| 7.42235 | 0.134174 | standard |
| 7.47849 | 0.224892 | standard |
| 7.53711 | 0.127724 | standard |
| 7.59881 | 0.143396 | standard |
| -0.898884 | 1.125e-06 | standard |
| 3.63378 | 1.0 | standard |
| 6.87557 | 0.046261 | standard |
| 6.96253 | 0.199126 | standard |
| 6.98091 | 0.176523 | standard |
| 7.01502 | 0.274406 | standard |
| 7.05081 | 0.232897 | standard |
| 7.07581 | 0.284617 | standard |
| 7.155 | 0.582451 | standard |
| 7.2459 | 0.0605654 | standard |
| 7.30194 | 0.233935 | standard |
| 7.36602 | 0.0945282 | standard |
| 7.39734 | 0.132456 | standard |
| 7.49132 | 0.215293 | standard |
| 7.55179 | 0.0989813 | standard |
| 7.58035 | 0.157722 | standard |
| -0.942625 | 1.125e-06 | standard |
| 3.63378 | 1.0 | standard |
| 6.88707 | 0.0495111 | standard |
| 6.96419 | 0.197061 | standard |
| 6.99342 | 0.168911 | standard |
| 7.01658 | 0.284389 | standard |
| 7.04312 | 0.211815 | standard |
| 7.07453 | 0.268707 | standard |
| 7.1542 | 0.586045 | standard |
| 7.25284 | 0.0489783 | standard |
| 7.30598 | 0.233958 | standard |
| 7.3897 | 0.14141 | standard |
| 7.441 | 0.0451253 | standard |
| 7.49561 | 0.217402 | standard |
| 7.57449 | 0.165302 | standard |
| -0.986763 | 1.125e-06 | standard |
| 3.63378 | 1.0 | standard |
| 6.89655 | 0.0535492 | standard |
| 6.96649 | 0.19722 | standard |
| 7.01771 | 0.293255 | standard |
| 7.03679 | 0.20802 | standard |
| 7.07318 | 0.25723 | standard |
| 7.15404 | 0.552421 | standard |
| 7.25777 | 0.040258 | standard |
| 7.30926 | 0.235517 | standard |
| 7.38417 | 0.147078 | standard |
| 7.44447 | 0.0353883 | standard |
| 7.49905 | 0.220552 | standard |
| 7.56993 | 0.172465 | standard |
| 7.61973 | 0.0181231 | standard |
| 3.63378 | 1.0 | standard |
| 6.94063 | 0.0815179 | standard |
| 6.97653 | 0.225126 | standard |
| 7.01226 | 0.205701 | standard |
| 7.03055 | 0.186373 | standard |
| 7.06757 | 0.238659 | standard |
| 7.10375 | 0.110039 | standard |
| 7.1545 | 0.503404 | standard |
| 7.26941 | 0.00740912 | standard |
| 7.32402 | 0.273036 | standard |
| 7.3614 | 0.209806 | standard |
| 7.51478 | 0.262213 | standard |
| 7.55054 | 0.231175 | standard |
| 3.63378 | 1.0 | standard |
| 6.95483 | 0.0950572 | standard |
| 6.97903 | 0.240521 | standard |
| 7.00331 | 0.177869 | standard |
| 7.04081 | 0.161614 | standard |
| 7.06592 | 0.240246 | standard |
| 7.09039 | 0.112953 | standard |
| 7.1545 | 0.50134 | standard |
| 7.32917 | 0.273521 | standard |
| 7.35472 | 0.228817 | standard |
| 7.52009 | 0.265324 | standard |
| 7.54451 | 0.243672 | standard |
| 3.63378 | 1.0 | standard |
| 6.96171 | 0.103104 | standard |
| 6.97992 | 0.245989 | standard |
| 6.99832 | 0.165606 | standard |
| 7.04636 | 0.152629 | standard |
| 7.06527 | 0.239642 | standard |
| 7.08376 | 0.116117 | standard |
| 7.1545 | 0.500728 | standard |
| 7.33198 | 0.270422 | standard |
| 7.35135 | 0.236328 | standard |
| 7.52291 | 0.263361 | standard |
| 7.54149 | 0.246892 | standard |
| 3.63378 | 1.0 | standard |
| 6.96572 | 0.109679 | standard |
| 6.98037 | 0.248159 | standard |
| 6.9951 | 0.156241 | standard |
| 7.04982 | 0.14765 | standard |
| 7.065 | 0.238818 | standard |
| 7.07985 | 0.118424 | standard |
| 7.1545 | 0.500505 | standard |
| 7.33379 | 0.267871 | standard |
| 7.34932 | 0.240165 | standard |
| 7.52472 | 0.258578 | standard |
| 7.53959 | 0.251944 | standard |
| 3.63378 | 1.0 | standard |
| 6.96839 | 0.112888 | standard |
| 6.98063 | 0.249945 | standard |
| 6.99287 | 0.151868 | standard |
| 7.05215 | 0.144515 | standard |
| 7.06478 | 0.238227 | standard |
| 7.07717 | 0.120117 | standard |
| 7.1545 | 0.500348 | standard |
| 7.33497 | 0.265681 | standard |
| 7.34796 | 0.242707 | standard |
| 7.52588 | 0.257914 | standard |
| 7.53832 | 0.252375 | standard |
| 3.63378 | 1.0 | standard |
| 6.97027 | 0.116373 | standard |
| 6.98076 | 0.250966 | standard |
| 6.99128 | 0.147294 | standard |
| 7.05384 | 0.142218 | standard |
| 7.06471 | 0.237016 | standard |
| 7.07534 | 0.121468 | standard |
| 7.1545 | 0.500231 | standard |
| 7.33587 | 0.264044 | standard |
| 7.34702 | 0.244571 | standard |
| 7.52675 | 0.255141 | standard |
| 7.53743 | 0.254941 | standard |
| 3.63378 | 1.0 | standard |
| 6.97102 | 0.117385 | standard |
| 6.98081 | 0.250595 | standard |
| 6.99064 | 0.146264 | standard |
| 7.05453 | 0.141396 | standard |
| 7.06464 | 0.23873 | standard |
| 7.0746 | 0.121849 | standard |
| 7.1545 | 0.500201 | standard |
| 7.33621 | 0.263694 | standard |
| 7.34662 | 0.245255 | standard |
| 7.5271 | 0.254843 | standard |
| 7.53706 | 0.255139 | standard |
| 3.63378 | 1.0 | standard |
| 6.97166 | 0.118011 | standard |
| 6.98086 | 0.250746 | standard |
| 6.99004 | 0.145284 | standard |
| 7.05512 | 0.140629 | standard |
| 7.06464 | 0.236854 | standard |
| 7.07395 | 0.122497 | standard |
| 7.1545 | 0.500176 | standard |
| 7.33651 | 0.263099 | standard |
| 7.34632 | 0.245799 | standard |
| 7.5274 | 0.254767 | standard |
| 7.53676 | 0.255106 | standard |
| 3.63378 | 1.0 | standard |
| 6.97275 | 0.119334 | standard |
| 6.98091 | 0.249881 | standard |
| 6.9891 | 0.143336 | standard |
| 7.05616 | 0.139249 | standard |
| 7.06457 | 0.237723 | standard |
| 7.07286 | 0.123322 | standard |
| 7.1545 | 0.500139 | standard |
| 7.33706 | 0.261787 | standard |
| 7.34573 | 0.246848 | standard |
| 7.52794 | 0.255162 | standard |
| 7.53622 | 0.254726 | standard |
| 3.63378 | 1.0 | standard |
| 6.9732 | 0.120084 | standard |
| 6.98094 | 0.251021 | standard |
| 6.9887 | 0.1426 | standard |
| 7.05661 | 0.138987 | standard |
| 7.06457 | 0.236002 | standard |
| 7.07247 | 0.123625 | standard |
| 7.1545 | 0.500125 | standard |
| 7.3373 | 0.261834 | standard |
| 7.34553 | 0.247253 | standard |
| 7.52814 | 0.255172 | standard |
| 7.53597 | 0.255171 | standard |
| 3.63378 | 1.0 | standard |
| 6.97359 | 0.120614 | standard |
| 6.98096 | 0.249638 | standard |
| 6.98836 | 0.142214 | standard |
| 7.05696 | 0.139982 | standard |
| 7.06455 | 0.237761 | standard |
| 7.07202 | 0.123731 | standard |
| 7.1545 | 0.500137 | standard |
| 7.3375 | 0.259768 | standard |
| 7.34533 | 0.249405 | standard |
| 7.52834 | 0.255177 | standard |
| 7.53577 | 0.2552 | standard |
| 3.63378 | 1.0 | standard |
| 6.97429 | 0.121474 | standard |
| 6.98102 | 0.252009 | standard |
| 6.98766 | 0.141306 | standard |
| 7.0576 | 0.139355 | standard |
| 7.06452 | 0.239632 | standard |
| 7.07128 | 0.123069 | standard |
| 7.1545 | 0.500134 | standard |
| 7.3378 | 0.257775 | standard |
| 7.34494 | 0.251471 | standard |
| 7.52863 | 0.255076 | standard |
| 7.53547 | 0.255073 | standard |
| 3.63378 | 1.0 | standard |
| 6.97538 | 0.122963 | standard |
| 6.98102 | 0.251244 | standard |
| 6.98667 | 0.139604 | standard |
| 7.05869 | 0.138075 | standard |
| 7.06446 | 0.241215 | standard |
| 7.07029 | 0.124005 | standard |
| 7.1545 | 0.500096 | standard |
| 7.33839 | 0.257433 | standard |
| 7.34434 | 0.2521 | standard |
| 7.52913 | 0.255046 | standard |
| 7.53493 | 0.254127 | standard |