4-Chlorophenoxyacetic acid
Simulation outputs:
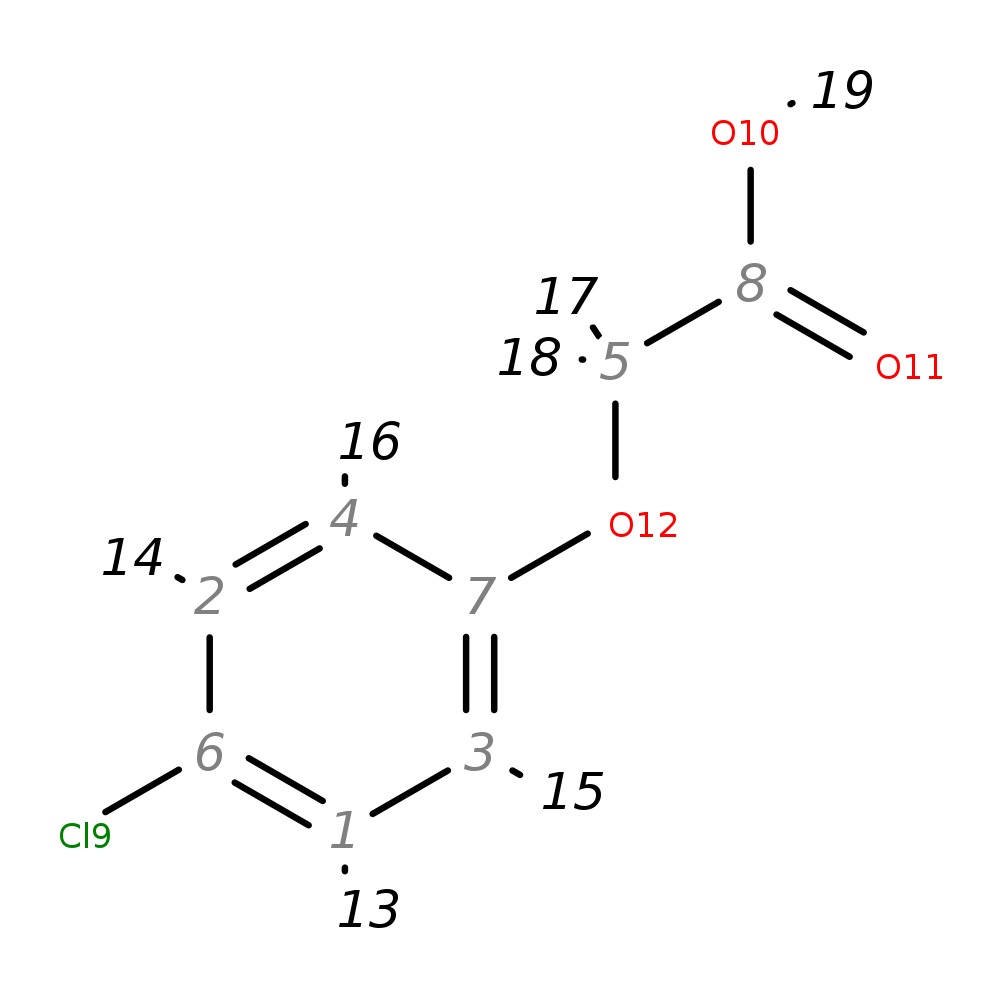
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.00727 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H7ClO3/c9-6-1-3-7(4-2-6)12-5-8(10)11/h1-4H,5H2,(H,10,11) | |
| Note 1 | Oscar Li |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4-chlorophenoxyacetic acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|
| 13 | 7.327 | 1.57 | 8.697 | 0.555 | 0 | 0 |
| 14 | 0 | 7.327 | 0.555 | 8.697 | 0 | 0 |
| 15 | 0 | 0 | 6.903 | 1.316 | 0 | 0 |
| 16 | 0 | 0 | 0 | 6.903 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 4.459 | -12.555 |
| 18 | 0 | 0 | 0 | 0 | 0 | 4.459 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 4.45881 | 1.0 | standard |
| 6.89347 | 0.374544 | standard |
| 6.91135 | 0.403762 | standard |
| 7.31806 | 0.40103 | standard |
| 7.33599 | 0.374526 | standard |
| -0.91148 | 3.125e-06 | standard |
| 4.45881 | 1.0 | standard |
| 6.76458 | 0.209695 | standard |
| 6.98998 | 0.593344 | standard |
| 7.23947 | 0.593329 | standard |
| 7.46487 | 0.209654 | standard |
| -0.99936 | 2.125e-06 | standard |
| 4.45881 | 1.0 | standard |
| 6.81625 | 0.261247 | standard |
| 6.96588 | 0.526653 | standard |
| 7.26358 | 0.526468 | standard |
| 7.41321 | 0.26105 | standard |
| -0.853159 | 1.125e-06 | standard |
| 4.45881 | 1.0 | standard |
| 6.83999 | 0.290491 | standard |
| 6.95206 | 0.492272 | standard |
| 7.27738 | 0.492566 | standard |
| 7.38941 | 0.290768 | standard |
| -0.915646 | 1.125e-06 | standard |
| 4.45881 | 1.0 | standard |
| 6.84763 | 0.301335 | standard |
| 6.94718 | 0.479168 | standard |
| 7.28227 | 0.479167 | standard |
| 7.38183 | 0.301172 | standard |
| -0.976447 | 1.125e-06 | standard |
| 4.45881 | 1.0 | standard |
| 6.8536 | 0.309766 | standard |
| 6.94314 | 0.470212 | standard |
| 7.2863 | 0.47035 | standard |
| 7.37586 | 0.309865 | standard |
| 4.45881 | 1.0 | standard |
| 6.87914 | 0.34869 | standard |
| 6.92392 | 0.428901 | standard |
| 7.30554 | 0.42855 | standard |
| 7.35031 | 0.347892 | standard |
| 4.45881 | 1.0 | standard |
| 6.88719 | 0.36144 | standard |
| 6.91701 | 0.416452 | standard |
| 7.3124 | 0.416401 | standard |
| 7.34227 | 0.361425 | standard |
| 4.45881 | 1.0 | standard |
| 6.89111 | 0.367716 | standard |
| 6.91351 | 0.410731 | standard |
| 7.3159 | 0.408958 | standard |
| 7.3383 | 0.369401 | standard |
| 4.45881 | 1.0 | standard |
| 6.89347 | 0.374515 | standard |
| 6.91135 | 0.403722 | standard |
| 7.31806 | 0.401774 | standard |
| 7.33599 | 0.374499 | standard |
| 4.45881 | 1.0 | standard |
| 6.89501 | 0.377493 | standard |
| 6.90996 | 0.396678 | standard |
| 7.31949 | 0.399389 | standard |
| 7.3344 | 0.378632 | standard |
| 4.45881 | 1.0 | standard |
| 6.89615 | 0.382246 | standard |
| 6.9089 | 0.396168 | standard |
| 7.32055 | 0.396167 | standard |
| 7.33331 | 0.382246 | standard |
| 4.45881 | 1.0 | standard |
| 6.89653 | 0.384302 | standard |
| 6.90852 | 0.393091 | standard |
| 7.32094 | 0.393105 | standard |
| 7.33293 | 0.381012 | standard |
| 4.45881 | 1.0 | standard |
| 6.89692 | 0.384073 | standard |
| 6.90814 | 0.391574 | standard |
| 7.32132 | 0.391574 | standard |
| 7.33253 | 0.384073 | standard |
| 4.45881 | 1.0 | standard |
| 6.89759 | 0.385766 | standard |
| 6.90751 | 0.392676 | standard |
| 7.3219 | 0.389549 | standard |
| 7.33187 | 0.383935 | standard |
| 4.45881 | 1.0 | standard |
| 6.89787 | 0.390887 | standard |
| 6.90726 | 0.389933 | standard |
| 7.32219 | 0.389933 | standard |
| 7.33159 | 0.390887 | standard |
| 4.45881 | 1.0 | standard |
| 6.89805 | 0.388021 | standard |
| 6.90698 | 0.396415 | standard |
| 7.32243 | 0.391707 | standard |
| 7.33139 | 0.384546 | standard |
| 4.45881 | 1.0 | standard |
| 6.89844 | 0.385691 | standard |
| 6.90659 | 0.389577 | standard |
| 7.32285 | 0.392891 | standard |
| 7.33091 | 0.389101 | standard |
| 4.45881 | 1.0 | standard |
| 6.89911 | 0.390421 | standard |
| 6.90603 | 0.390936 | standard |
| 7.32343 | 0.39098 | standard |
| 7.33033 | 0.380308 | standard |