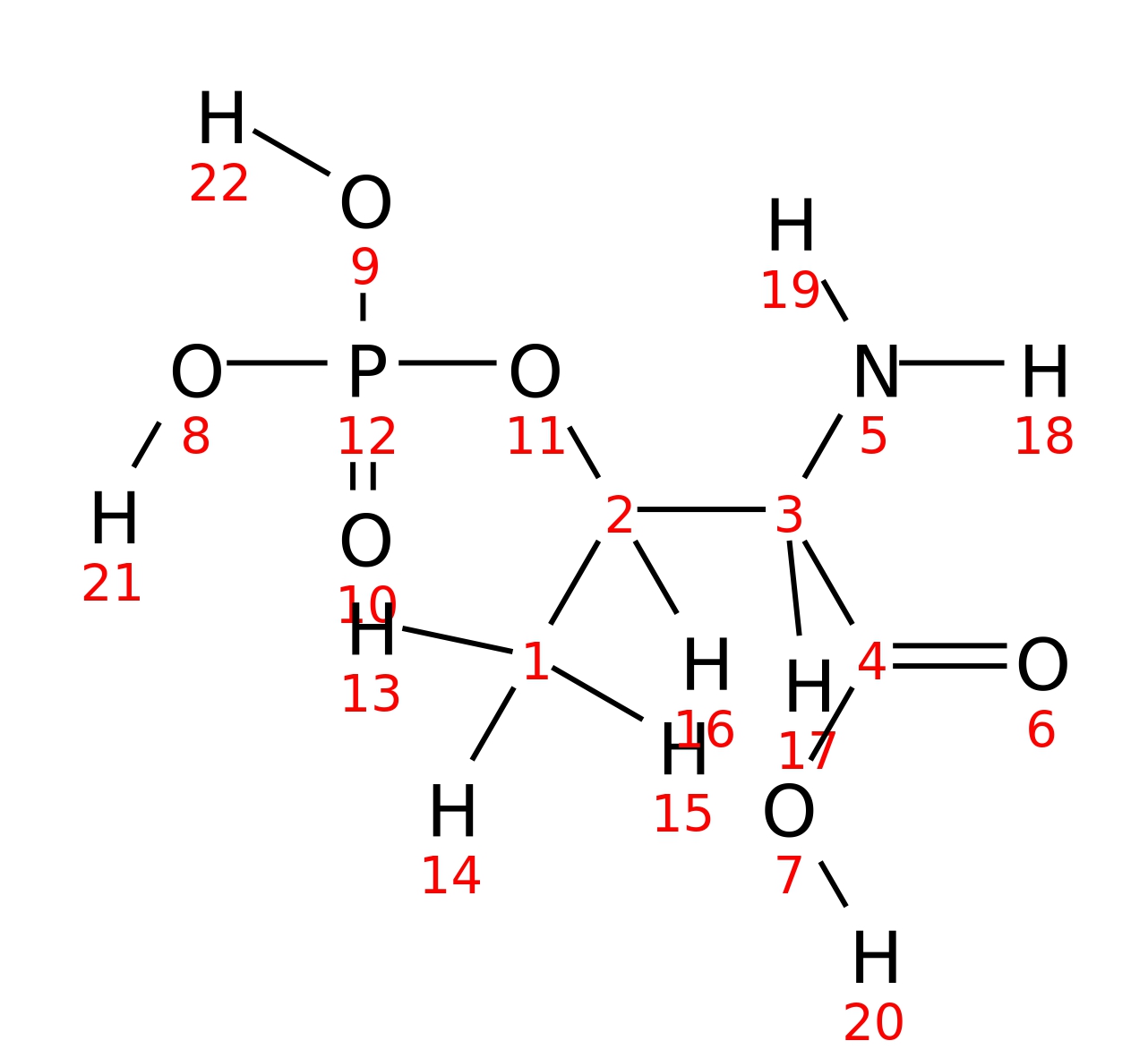
O-phospho-DL-threonine
Simulation outputs:
|
Parameter | Value | |||
| Field strength | 499.84(MHz) | ||||
| RMSD of the fit | 0.01313 | ||||
| Temperature | 298 K | ||||
| pH | 7.4 | ||||
| InChI | InChI=1S/C4H10NO6P/c1-2(3(5)4(6)7)11-12(8,9)10/h2-3H,5H2,1H3,(H,6,7)(H2,8,9,10)/t2-,3+/m1/s1 | ||||
| Note 1 | 31P coupling | ||||
| Additional coupling constants |
|
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| O-phospho-DL-threonine | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|
| 13 | 1.378 | -12.5 | -12.5 | 6.274 | 0 |
| 14 | 0 | 1.378 | -12.5 | 6.274 | 0 |
| 15 | 0 | 0 | 1.378 | 6.274 | 0 |
| 16 | 0 | 0 | 0 | 4.33 | 8.03 |
| 17 | 0 | 0 | 0 | 0 | 3.547 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.37204 | 1.00001 | standard |
| 1.38455 | 0.999269 | standard |
| 3.53878 | 0.434573 | standard |
| 3.55483 | 0.436894 | standard |
| 4.29542 | 0.0321402 | standard |
| 4.30807 | 0.0999721 | standard |
| 4.31131 | 0.0829951 | standard |
| 4.32409 | 0.194335 | standard |
| 4.33681 | 0.194679 | standard |
| 4.34964 | 0.0832938 | standard |
| 4.35283 | 0.100097 | standard |
| 4.36551 | 0.0315499 | standard |
| 1.29939 | 0.951048 | standard |
| 1.45316 | 1.0 | standard |
| 3.28013 | 0.00754413 | standard |
| 3.43409 | 0.331732 | standard |
| 3.63416 | 0.428389 | standard |
| 3.72608 | 0.0558841 | standard |
| 3.91681 | 0.0377761 | standard |
| 4.07045 | 0.0943221 | standard |
| 4.11712 | 0.079646 | standard |
| 4.27084 | 0.15896 | standard |
| 4.42703 | 0.146163 | standard |
| 4.58533 | 0.0605001 | standard |
| 4.6257 | 0.0682453 | standard |
| 4.7835 | 0.0222351 | standard |
| 1.3256 | 0.967621 | standard |
| 1.42917 | 1.0 | standard |
| 3.3813 | 0.00934212 | standard |
| 3.47441 | 0.367703 | standard |
| 3.60802 | 0.435661 | standard |
| 4.04856 | 0.033565 | standard |
| 4.1528 | 0.0965151 | standard |
| 4.18134 | 0.0804542 | standard |
| 4.28631 | 0.17186 | standard |
| 4.39123 | 0.162829 | standard |
| 4.49725 | 0.0673261 | standard |
| 4.52413 | 0.0780794 | standard |
| 4.62951 | 0.024873 | standard |
| 1.33881 | 0.978233 | standard |
| 1.41676 | 1.0 | standard |
| 3.42911 | 0.0109441 | standard |
| 3.49358 | 0.386031 | standard |
| 3.59386 | 0.438359 | standard |
| 4.11692 | 0.032864 | standard |
| 4.19552 | 0.097592 | standard |
| 4.21635 | 0.0810946 | standard |
| 4.29566 | 0.177801 | standard |
| 4.37458 | 0.172323 | standard |
| 4.4543 | 0.0714064 | standard |
| 4.47442 | 0.0841284 | standard |
| 4.55349 | 0.0265494 | standard |
| 1.34321 | 0.982116 | standard |
| 1.41259 | 1.0 | standard |
| 3.49981 | 0.392199 | standard |
| 3.58896 | 0.439068 | standard |
| 4.14003 | 0.03262 | standard |
| 4.21004 | 0.0976551 | standard |
| 4.2284 | 0.081305 | standard |
| 4.29904 | 0.179819 | standard |
| 4.36921 | 0.175315 | standard |
| 4.44018 | 0.072831 | standard |
| 4.45796 | 0.0861937 | standard |
| 4.52826 | 0.027111 | standard |
| 1.34674 | 0.983833 | standard |
| 1.40922 | 1.0 | standard |
| 3.50475 | 0.397088 | standard |
| 3.58497 | 0.439483 | standard |
| 4.15864 | 0.0325283 | standard |
| 4.22169 | 0.0980815 | standard |
| 4.23816 | 0.0815593 | standard |
| 4.30181 | 0.181604 | standard |
| 4.36498 | 0.177735 | standard |
| 4.42886 | 0.073853 | standard |
| 4.44493 | 0.087636 | standard |
| 4.50816 | 0.0275676 | standard |
| 1.36255 | 0.999785 | standard |
| 1.39387 | 1.00001 | standard |
| 3.52636 | 0.417819 | standard |
| 3.56648 | 0.439832 | standard |
| 4.24361 | 0.0322541 | standard |
| 4.2752 | 0.100719 | standard |
| 4.28334 | 0.0829741 | standard |
| 4.31526 | 0.191535 | standard |
| 4.34691 | 0.190154 | standard |
| 4.37892 | 0.0808061 | standard |
| 4.38693 | 0.0959681 | standard |
| 4.41862 | 0.0305598 | standard |
| 1.36781 | 0.999915 | standard |
| 1.3887 | 1.0 | standard |
| 3.53331 | 0.427931 | standard |
| 3.56008 | 0.437645 | standard |
| 4.27233 | 0.0321804 | standard |
| 4.29341 | 0.100252 | standard |
| 4.29881 | 0.0829969 | standard |
| 4.32012 | 0.193113 | standard |
| 4.34122 | 0.192297 | standard |
| 4.36254 | 0.0824441 | standard |
| 4.36787 | 0.0984143 | standard |
| 4.389 | 0.0311095 | standard |
| 1.37045 | 1.0 | standard |
| 1.38611 | 1.00001 | standard |
| 3.53678 | 0.432496 | standard |
| 3.55681 | 0.436218 | standard |
| 4.28674 | 0.032144 | standard |
| 4.30259 | 0.100347 | standard |
| 4.30655 | 0.0832776 | standard |
| 4.32259 | 0.193459 | standard |
| 4.33844 | 0.19338 | standard |
| 4.35441 | 0.0828506 | standard |
| 4.35844 | 0.098937 | standard |
| 4.37429 | 0.0313579 | standard |
| 1.37204 | 1.00001 | standard |
| 1.38455 | 0.998834 | standard |
| 3.53885 | 0.434401 | standard |
| 3.55483 | 0.435792 | standard |
| 4.29543 | 0.03212 | standard |
| 4.30807 | 0.099924 | standard |
| 4.31131 | 0.0829671 | standard |
| 4.32409 | 0.194281 | standard |
| 4.3368 | 0.193902 | standard |
| 4.34959 | 0.082831 | standard |
| 4.35279 | 0.099586 | standard |
| 4.36541 | 0.0315558 | standard |
| 1.37306 | 0.999363 | standard |
| 1.38354 | 1.00001 | standard |
| 3.54015 | 0.44041 | standard |
| 3.55354 | 0.44041 | standard |
| 4.30125 | 0.0321717 | standard |
| 4.3118 | 0.10103 | standard |
| 4.31449 | 0.0840559 | standard |
| 4.32513 | 0.195744 | standard |
| 4.33566 | 0.196261 | standard |
| 4.34636 | 0.0843233 | standard |
| 4.349 | 0.101237 | standard |
| 4.35959 | 0.0321507 | standard |
| 1.37383 | 1.00001 | standard |
| 1.38274 | 1.00001 | standard |
| 3.54115 | 0.439731 | standard |
| 3.55265 | 0.440216 | standard |
| 4.3054 | 0.032177 | standard |
| 4.31441 | 0.101118 | standard |
| 4.31669 | 0.0838111 | standard |
| 4.32586 | 0.196542 | standard |
| 4.33489 | 0.19591 | standard |
| 4.34404 | 0.0838634 | standard |
| 4.34633 | 0.101143 | standard |
| 4.35534 | 0.0321808 | standard |
| 1.37413 | 1.00001 | standard |
| 1.38245 | 1.00001 | standard |
| 3.54154 | 0.440252 | standard |
| 3.55225 | 0.440252 | standard |
| 4.30707 | 0.0321744 | standard |
| 4.31548 | 0.100829 | standard |
| 4.31767 | 0.0837011 | standard |
| 4.32616 | 0.195799 | standard |
| 4.33457 | 0.196189 | standard |
| 4.3432 | 0.0848934 | standard |
| 4.34525 | 0.10168 | standard |
| 4.35376 | 0.032109 | standard |
| 1.37442 | 0.99971 | standard |
| 1.38225 | 1.00001 | standard |
| 3.54184 | 0.440223 | standard |
| 3.55195 | 0.440223 | standard |
| 4.30851 | 0.0321565 | standard |
| 4.31642 | 0.101094 | standard |
| 4.31841 | 0.0840254 | standard |
| 4.32641 | 0.195524 | standard |
| 4.33433 | 0.19601 | standard |
| 4.34232 | 0.0840355 | standard |
| 4.34432 | 0.101169 | standard |
| 4.35223 | 0.0321575 | standard |
| 1.37482 | 1.00001 | standard |
| 1.38175 | 1.00001 | standard |
| 3.54243 | 0.440246 | standard |
| 3.55136 | 0.440247 | standard |
| 4.31086 | 0.0320952 | standard |
| 4.31797 | 0.101859 | standard |
| 4.31965 | 0.0851473 | standard |
| 4.32687 | 0.19626 | standard |
| 4.33387 | 0.195787 | standard |
| 4.34095 | 0.0836916 | standard |
| 4.34277 | 0.100825 | standard |
| 4.34978 | 0.0321729 | standard |
| 1.37502 | 1.00002 | standard |
| 1.38158 | 0.998223 | standard |
| 3.54263 | 0.440013 | standard |
| 3.55116 | 0.440149 | standard |
| 4.31189 | 0.0321077 | standard |
| 4.3186 | 0.101115 | standard |
| 4.32029 | 0.0843583 | standard |
| 4.32701 | 0.195415 | standard |
| 4.33369 | 0.196824 | standard |
| 4.3404 | 0.0832075 | standard |
| 4.34214 | 0.100603 | standard |
| 4.34875 | 0.0321917 | standard |
| 1.37519 | 0.998358 | standard |
| 1.38146 | 1.00001 | standard |
| 3.54293 | 0.440374 | standard |
| 3.55096 | 0.439749 | standard |
| 4.3128 | 0.0320951 | standard |
| 4.31921 | 0.101994 | standard |
| 4.32071 | 0.0853587 | standard |
| 4.32722 | 0.196354 | standard |
| 4.33352 | 0.195516 | standard |
| 4.33989 | 0.0836372 | standard |
| 4.34153 | 0.10054 | standard |
| 4.34784 | 0.0321727 | standard |
| 1.37541 | 1.00001 | standard |
| 1.38116 | 0.999703 | standard |
| 3.54323 | 0.440443 | standard |
| 3.55056 | 0.440443 | standard |
| 4.31446 | 0.0322138 | standard |
| 4.32017 | 0.100929 | standard |
| 4.32164 | 0.0833491 | standard |
| 4.32746 | 0.197618 | standard |
| 4.33322 | 0.194861 | standard |
| 4.33905 | 0.084887 | standard |
| 4.34047 | 0.101585 | standard |
| 4.34628 | 0.0321224 | standard |
| 1.37591 | 1.00001 | standard |
| 1.38074 | 0.996899 | standard |
| 3.54382 | 0.44042 | standard |
| 3.54997 | 0.440235 | standard |
| 4.31691 | 0.0322262 | standard |
| 4.32172 | 0.100271 | standard |
| 4.32301 | 0.0828333 | standard |
| 4.32788 | 0.19641 | standard |
| 4.33277 | 0.196433 | standard |
| 4.33776 | 0.085014 | standard |
| 4.33891 | 0.101899 | standard |
| 4.34382 | 0.0321221 | standard |