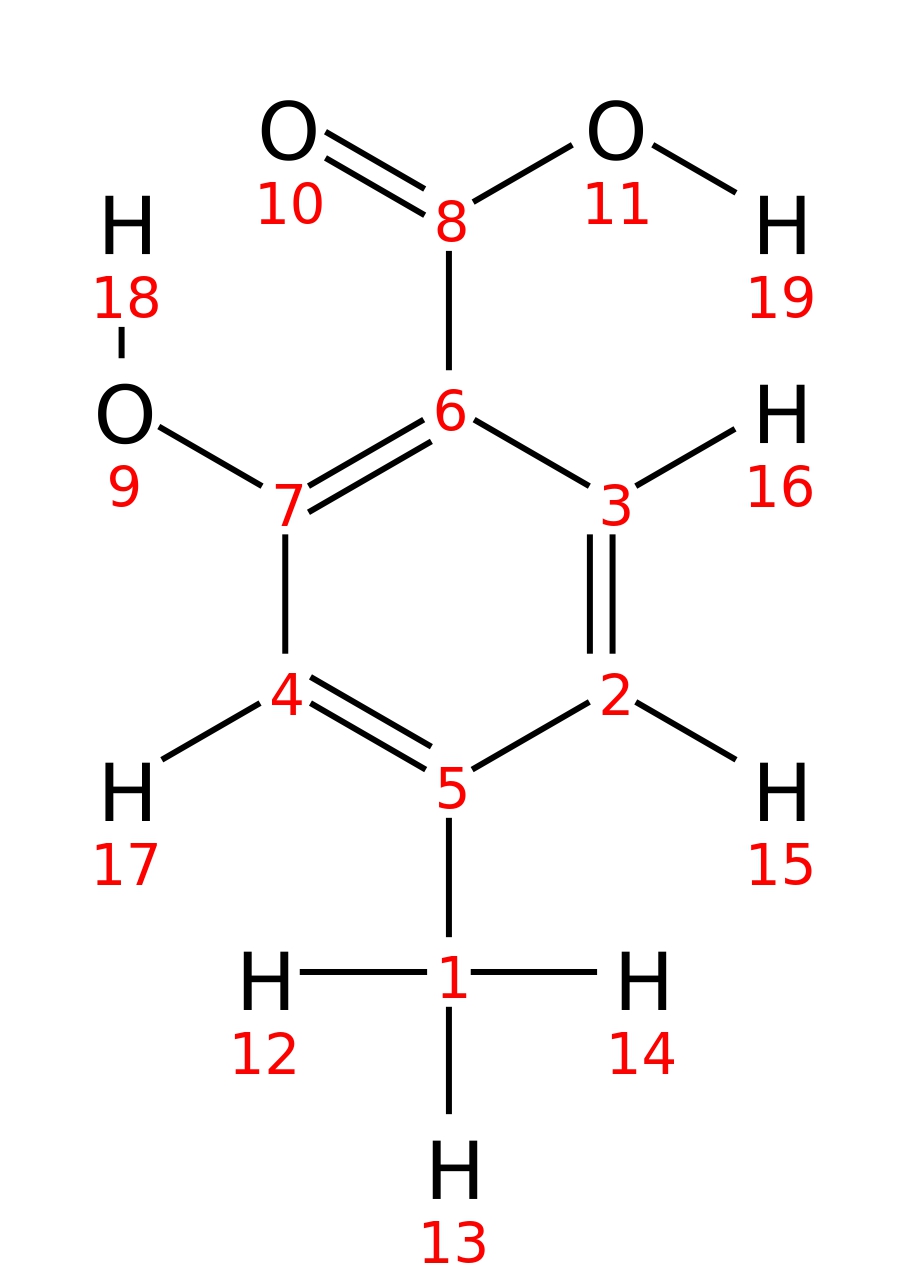
4-Methylsalicylic-acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01297 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H8O3/c1-5-2-3-6(8(10)11)7(9)4-5/h2-4,9H,1H3,(H,10,11) | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4-methylsalicylic acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 12 | 13 | 14 | 17 | 16 | 15 | |
|---|---|---|---|---|---|---|
| 12 | 2.309 | -14.9 | -14.9 | 0 | 0 | 0 |
| 13 | 0 | 2.31 | -14.9 | 0 | 0 | 0 |
| 14 | 0 | 0 | 2.31 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 6.763 | 0 | 1.746 |
| 16 | 0 | 0 | 0 | 0 | 7.686 | 7.952 |
| 15 | 0 | 0 | 0 | 0 | 0 | 6.789 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.30955 | 1.0 | standard |
| 6.76119 | 0.196162 | standard |
| 6.76407 | 0.23095 | standard |
| 6.77934 | 0.123793 | standard |
| 6.7822 | 0.101401 | standard |
| 6.79514 | 0.115806 | standard |
| 6.79806 | 0.102631 | standard |
| 7.67796 | 0.16901 | standard |
| 7.69382 | 0.16901 | standard |
| 2.30955 | 1.0 | standard |
| 6.70048 | 0.162828 | standard |
| 6.74696 | 0.243271 | standard |
| 6.77794 | 0.237005 | standard |
| 6.85981 | 0.176489 | standard |
| 7.59319 | 0.185689 | standard |
| 7.68401 | 0.0387261 | standard |
| 7.80006 | 0.119363 | standard |
| 2.30955 | 1.0 | standard |
| 6.7507 | 0.267294 | standard |
| 6.83844 | 0.159572 | standard |
| 7.62157 | 0.172387 | standard |
| 7.71466 | 0.0361371 | standard |
| 7.75971 | 0.127425 | standard |
| 2.30955 | 1.0 | standard |
| 6.75214 | 0.308115 | standard |
| 6.82688 | 0.150084 | standard |
| 7.63661 | 0.163258 | standard |
| 7.74044 | 0.13002 | standard |
| 2.30955 | 1.0 | standard |
| 6.75335 | 0.334051 | standard |
| 6.82292 | 0.146741 | standard |
| 7.64175 | 0.158959 | standard |
| 7.7341 | 0.129906 | standard |
| 2.30955 | 1.0 | standard |
| 6.7552 | 0.358223 | standard |
| 6.81963 | 0.143844 | standard |
| 7.64594 | 0.155031 | standard |
| 7.72904 | 0.12938 | standard |
| 2.30955 | 1.0 | standard |
| 6.76566 | 0.413243 | standard |
| 6.80463 | 0.128492 | standard |
| 6.81181 | 0.10385 | standard |
| 7.66721 | 0.159319 | standard |
| 7.70525 | 0.148326 | standard |
| 2.30955 | 1.0 | standard |
| 6.76496 | 0.264428 | standard |
| 6.77276 | 0.169367 | standard |
| 6.79938 | 0.121657 | standard |
| 6.80431 | 0.102575 | standard |
| 7.67295 | 0.169913 | standard |
| 7.69903 | 0.163821 | standard |
| 2.30955 | 1.0 | standard |
| 6.76085 | 0.19257 | standard |
| 6.7644 | 0.239174 | standard |
| 6.77692 | 0.13327 | standard |
| 6.79676 | 0.117788 | standard |
| 6.80044 | 0.102255 | standard |
| 7.67603 | 0.169447 | standard |
| 7.69576 | 0.167678 | standard |
| 2.30955 | 1.0 | standard |
| 6.76119 | 0.196137 | standard |
| 6.76407 | 0.230922 | standard |
| 6.77934 | 0.123775 | standard |
| 6.7822 | 0.101386 | standard |
| 6.79514 | 0.115792 | standard |
| 6.79806 | 0.102616 | standard |
| 7.67796 | 0.169024 | standard |
| 7.69377 | 0.169128 | standard |
| 2.30955 | 1.0 | standard |
| 6.76137 | 0.197315 | standard |
| 6.76385 | 0.225777 | standard |
| 6.78081 | 0.118337 | standard |
| 6.78335 | 0.101034 | standard |
| 6.79412 | 0.114804 | standard |
| 6.79654 | 0.103352 | standard |
| 7.67925 | 0.169064 | standard |
| 7.69243 | 0.169064 | standard |
| 2.30955 | 1.0 | standard |
| 6.76157 | 0.2 | standard |
| 6.76366 | 0.223599 | standard |
| 6.782 | 0.116563 | standard |
| 6.78409 | 0.103057 | standard |
| 6.79331 | 0.11384 | standard |
| 6.79541 | 0.103558 | standard |
| 7.68019 | 0.169248 | standard |
| 7.69149 | 0.169248 | standard |
| 2.30955 | 1.0 | standard |
| 6.76166 | 0.201874 | standard |
| 6.76356 | 0.223465 | standard |
| 6.78249 | 0.116102 | standard |
| 6.7844 | 0.104008 | standard |
| 6.79299 | 0.112622 | standard |
| 6.79504 | 0.102602 | standard |
| 7.68054 | 0.16902 | standard |
| 7.6911 | 0.169253 | standard |
| 2.30955 | 1.0 | standard |
| 6.76168 | 0.200658 | standard |
| 6.76352 | 0.22108 | standard |
| 6.78288 | 0.114703 | standard |
| 6.78473 | 0.103449 | standard |
| 6.7928 | 0.113015 | standard |
| 6.79465 | 0.103663 | standard |
| 7.68089 | 0.169252 | standard |
| 7.6908 | 0.169252 | standard |
| 2.30955 | 1.0 | standard |
| 6.7618 | 0.202066 | standard |
| 6.76344 | 0.219695 | standard |
| 6.78346 | 0.112837 | standard |
| 6.78517 | 0.103088 | standard |
| 6.79228 | 0.11181 | standard |
| 6.79399 | 0.103065 | standard |
| 7.68138 | 0.169254 | standard |
| 7.6902 | 0.169254 | standard |
| 2.30955 | 1.0 | standard |
| 6.76178 | 0.200188 | standard |
| 6.7634 | 0.217642 | standard |
| 6.78375 | 0.112302 | standard |
| 6.78538 | 0.103263 | standard |
| 6.79208 | 0.111537 | standard |
| 6.79371 | 0.103104 | standard |
| 7.68168 | 0.169276 | standard |
| 7.69001 | 0.169276 | standard |
| 2.30955 | 1.0 | standard |
| 6.76188 | 0.203076 | standard |
| 6.76335 | 0.218691 | standard |
| 6.78406 | 0.113868 | standard |
| 6.78546 | 0.105765 | standard |
| 6.79199 | 0.113256 | standard |
| 6.79339 | 0.105613 | standard |
| 7.68188 | 0.169265 | standard |
| 7.68981 | 0.169265 | standard |
| 2.30955 | 1.0 | standard |
| 6.76196 | 0.203263 | standard |
| 6.76331 | 0.217862 | standard |
| 6.78444 | 0.112043 | standard |
| 6.78579 | 0.104741 | standard |
| 6.79168 | 0.111756 | standard |
| 6.79303 | 0.104436 | standard |
| 7.68217 | 0.169249 | standard |
| 7.68941 | 0.169249 | standard |
| 2.30955 | 1.0 | standard |
| 6.76203 | 0.201956 | standard |
| 6.76323 | 0.214093 | standard |
| 6.78513 | 0.112552 | standard |
| 6.7862 | 0.106922 | standard |
| 6.79116 | 0.109937 | standard |
| 6.79236 | 0.103743 | standard |
| 7.68277 | 0.169227 | standard |
| 7.68892 | 0.169227 | standard |