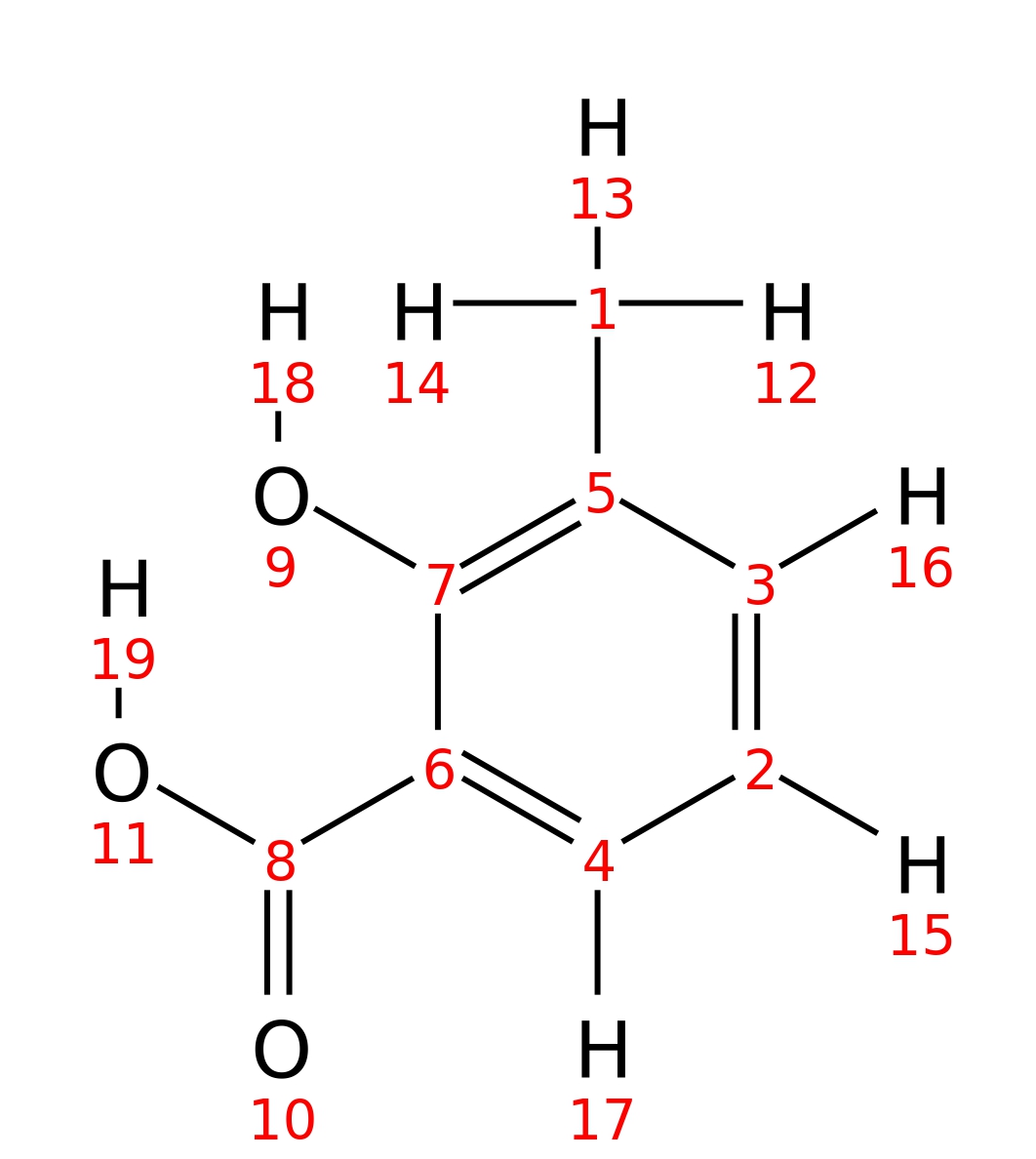
3-Methylsalicylic-acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.00959 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H8O3/c1-5-3-2-4-6(7(5)9)8(10)11/h2-4,9H,1H3,(H,10,11) | |
| Note 1 | 16?17 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-methylsalicylic acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 12 | 13 | 14 | 16 | 15 | 17 | |
|---|---|---|---|---|---|---|
| 12 | 2.207 | -14.9 | -14.9 | 1.0 | 0 | 0 |
| 13 | 0 | 2.207 | -14.9 | 1.0 | 0 | 0 |
| 14 | 0 | 0 | 2.207 | 1.0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 7.321 | 7.687 | 1.397 |
| 15 | 0 | 0 | 0 | 0 | 6.858 | 7.713 |
| 17 | 0 | 0 | 0 | 0 | 0 | 7.663 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.20683 | 1.00007 | standard |
| 2.20754 | 1.00007 | standard |
| 6.84286 | 0.114187 | standard |
| 6.85828 | 0.229033 | standard |
| 6.8737 | 0.119219 | standard |
| 7.31298 | 0.130158 | standard |
| 7.32841 | 0.123993 | standard |
| 7.65443 | 0.146669 | standard |
| 7.65676 | 0.147036 | standard |
| 7.66978 | 0.146167 | standard |
| 7.67224 | 0.14534 | standard |
| 2.20495 | 1.0 | standard |
| 2.20932 | 0.999976 | standard |
| 6.64017 | 0.0608224 | standard |
| 6.82891 | 0.188256 | standard |
| 7.01558 | 0.200189 | standard |
| 7.25284 | 0.168027 | standard |
| 7.42716 | 0.081167 | standard |
| 7.56294 | 0.185172 | standard |
| 7.60424 | 0.149683 | standard |
| 7.74653 | 0.113041 | standard |
| 7.788 | 0.102609 | standard |
| 2.20453 | 1.0 | standard |
| 2.2098 | 0.999916 | standard |
| 6.71783 | 0.0755299 | standard |
| 6.84482 | 0.212237 | standard |
| 6.97145 | 0.171805 | standard |
| 7.26785 | 0.156182 | standard |
| 7.3919 | 0.0918042 | standard |
| 7.594 | 0.171535 | standard |
| 7.6175 | 0.153942 | standard |
| 7.71956 | 0.12372 | standard |
| 7.74321 | 0.116128 | standard |
| 2.20496 | 0.999998 | standard |
| 2.20944 | 1.0 | standard |
| 6.75507 | 0.0844591 | standard |
| 6.85069 | 0.22085 | standard |
| 6.94624 | 0.15788 | standard |
| 7.27839 | 0.150704 | standard |
| 7.37301 | 0.101233 | standard |
| 7.61042 | 0.165895 | standard |
| 7.6269 | 0.154907 | standard |
| 7.70556 | 0.129932 | standard |
| 7.72206 | 0.124868 | standard |
| 2.20517 | 1.0 | standard |
| 2.2092 | 0.99999 | standard |
| 6.76721 | 0.0875821 | standard |
| 6.85231 | 0.222892 | standard |
| 6.93732 | 0.15298 | standard |
| 7.28243 | 0.148363 | standard |
| 7.36679 | 0.104185 | standard |
| 7.61608 | 0.163504 | standard |
| 7.63036 | 0.154782 | standard |
| 7.70084 | 0.13196 | standard |
| 7.71514 | 0.127512 | standard |
| 2.20534 | 1.0 | standard |
| 2.20908 | 0.999905 | standard |
| 6.77683 | 0.0902479 | standard |
| 6.85346 | 0.224292 | standard |
| 6.93008 | 0.149338 | standard |
| 7.2858 | 0.147054 | standard |
| 7.36189 | 0.106531 | standard |
| 7.62057 | 0.162591 | standard |
| 7.63328 | 0.154695 | standard |
| 7.69704 | 0.13376 | standard |
| 7.70967 | 0.12975 | standard |
| 2.20628 | 0.999877 | standard |
| 2.20811 | 1.0 | standard |
| 6.81877 | 0.103012 | standard |
| 6.85721 | 0.22789 | standard |
| 6.89563 | 0.1321 | standard |
| 7.30214 | 0.137654 | standard |
| 7.34062 | 0.117427 | standard |
| 7.64144 | 0.151913 | standard |
| 7.64754 | 0.152152 | standard |
| 7.6799 | 0.140433 | standard |
| 7.686 | 0.139166 | standard |
| 2.20646 | 1.00002 | standard |
| 2.20791 | 1.00002 | standard |
| 6.83225 | 0.109557 | standard |
| 6.85788 | 0.231801 | standard |
| 6.88352 | 0.126784 | standard |
| 7.30816 | 0.136327 | standard |
| 7.33375 | 0.123114 | standard |
| 7.64863 | 0.151239 | standard |
| 7.65256 | 0.152122 | standard |
| 7.67429 | 0.146241 | standard |
| 7.67822 | 0.145195 | standard |
| 2.20679 | 0.9997 | standard |
| 2.20769 | 1.0 | standard |
| 6.8389 | 0.111698 | standard |
| 6.85814 | 0.229665 | standard |
| 6.87737 | 0.121871 | standard |
| 7.31122 | 0.132548 | standard |
| 7.33039 | 0.122418 | standard |
| 7.65219 | 0.145539 | standard |
| 7.65524 | 0.147162 | standard |
| 7.67154 | 0.147157 | standard |
| 7.67444 | 0.14634 | standard |
| 2.20683 | 1.00006 | standard |
| 2.20755 | 1.00005 | standard |
| 6.84286 | 0.113734 | standard |
| 6.85828 | 0.229855 | standard |
| 6.8737 | 0.119202 | standard |
| 7.31301 | 0.129562 | standard |
| 7.32835 | 0.124751 | standard |
| 7.65443 | 0.147033 | standard |
| 7.65676 | 0.147625 | standard |
| 7.66978 | 0.14617 | standard |
| 7.67224 | 0.145294 | standard |
| 2.20688 | 0.999896 | standard |
| 2.20759 | 1.0 | standard |
| 6.84554 | 0.116515 | standard |
| 6.85833 | 0.231203 | standard |
| 6.87112 | 0.119617 | standard |
| 7.3143 | 0.13268 | standard |
| 7.32714 | 0.128047 | standard |
| 7.65587 | 0.14678 | standard |
| 7.6579 | 0.147831 | standard |
| 7.66878 | 0.150066 | standard |
| 7.67066 | 0.149296 | standard |
| 2.20701 | 1.00002 | standard |
| 2.20736 | 1.00001 | standard |
| 6.84732 | 0.114856 | standard |
| 6.85838 | 0.227459 | standard |
| 6.86934 | 0.116235 | standard |
| 7.31519 | 0.127097 | standard |
| 7.32621 | 0.125961 | standard |
| 7.65695 | 0.145657 | standard |
| 7.65861 | 0.146435 | standard |
| 7.66795 | 0.146435 | standard |
| 7.66961 | 0.145656 | standard |
| 2.2071 | 0.999997 | standard |
| 2.20737 | 1.0 | standard |
| 6.84812 | 0.114608 | standard |
| 6.85838 | 0.226872 | standard |
| 6.86864 | 0.115883 | standard |
| 7.31554 | 0.127307 | standard |
| 7.32579 | 0.125209 | standard |
| 7.65732 | 0.144412 | standard |
| 7.65892 | 0.145207 | standard |
| 7.66763 | 0.145144 | standard |
| 7.66923 | 0.144395 | standard |
| 2.20693 | 1.00039 | standard |
| 2.20754 | 1.00036 | standard |
| 6.84881 | 0.118108 | standard |
| 6.85838 | 0.233757 | standard |
| 6.86805 | 0.119491 | standard |
| 7.31581 | 0.131483 | standard |
| 7.32541 | 0.131094 | standard |
| 7.65771 | 0.148306 | standard |
| 7.65923 | 0.149284 | standard |
| 7.66732 | 0.14933 | standard |
| 7.66885 | 0.148481 | standard |
| 2.20708 | 1.00001 | standard |
| 2.20738 | 0.999812 | standard |
| 6.8499 | 0.116196 | standard |
| 6.85843 | 0.228794 | standard |
| 6.86696 | 0.116196 | standard |
| 7.31638 | 0.129707 | standard |
| 7.32488 | 0.128638 | standard |
| 7.65829 | 0.144755 | standard |
| 7.65964 | 0.144796 | standard |
| 7.66693 | 0.14913 | standard |
| 7.66814 | 0.148402 | standard |
| 2.20698 | 1.0 | standard |
| 2.20749 | 1.00001 | standard |
| 6.8503 | 0.119123 | standard |
| 6.85843 | 0.234988 | standard |
| 6.86656 | 0.119123 | standard |
| 7.31655 | 0.132302 | standard |
| 7.32471 | 0.130906 | standard |
| 7.65859 | 0.14929 | standard |
| 7.65984 | 0.150114 | standard |
| 7.66671 | 0.150114 | standard |
| 7.66797 | 0.149291 | standard |
| 2.20702 | 1.00021 | standard |
| 2.20735 | 1.00011 | standard |
| 6.8507 | 0.115776 | standard |
| 6.85843 | 0.229799 | standard |
| 6.86616 | 0.115806 | standard |
| 7.31678 | 0.12686 | standard |
| 7.3245 | 0.128913 | standard |
| 7.65879 | 0.146322 | standard |
| 7.65994 | 0.147868 | standard |
| 7.66651 | 0.147986 | standard |
| 7.66767 | 0.147347 | standard |
| 2.20712 | 1.00014 | standard |
| 2.20736 | 1.00014 | standard |
| 6.85139 | 0.116073 | standard |
| 6.85843 | 0.229012 | standard |
| 6.86547 | 0.116072 | standard |
| 7.31719 | 0.126676 | standard |
| 7.32409 | 0.126676 | standard |
| 7.65917 | 0.145286 | standard |
| 7.66025 | 0.146088 | standard |
| 7.6662 | 0.146088 | standard |
| 7.66729 | 0.145286 | standard |
| 2.20701 | 1.00033 | standard |
| 2.20736 | 1.00033 | standard |
| 6.85248 | 0.118836 | standard |
| 6.85843 | 0.234455 | standard |
| 6.86438 | 0.118837 | standard |
| 7.31775 | 0.133168 | standard |
| 7.32355 | 0.130877 | standard |
| 7.65985 | 0.148732 | standard |
| 7.66076 | 0.14957 | standard |
| 7.66582 | 0.154712 | standard |
| 7.66658 | 0.154011 | standard |