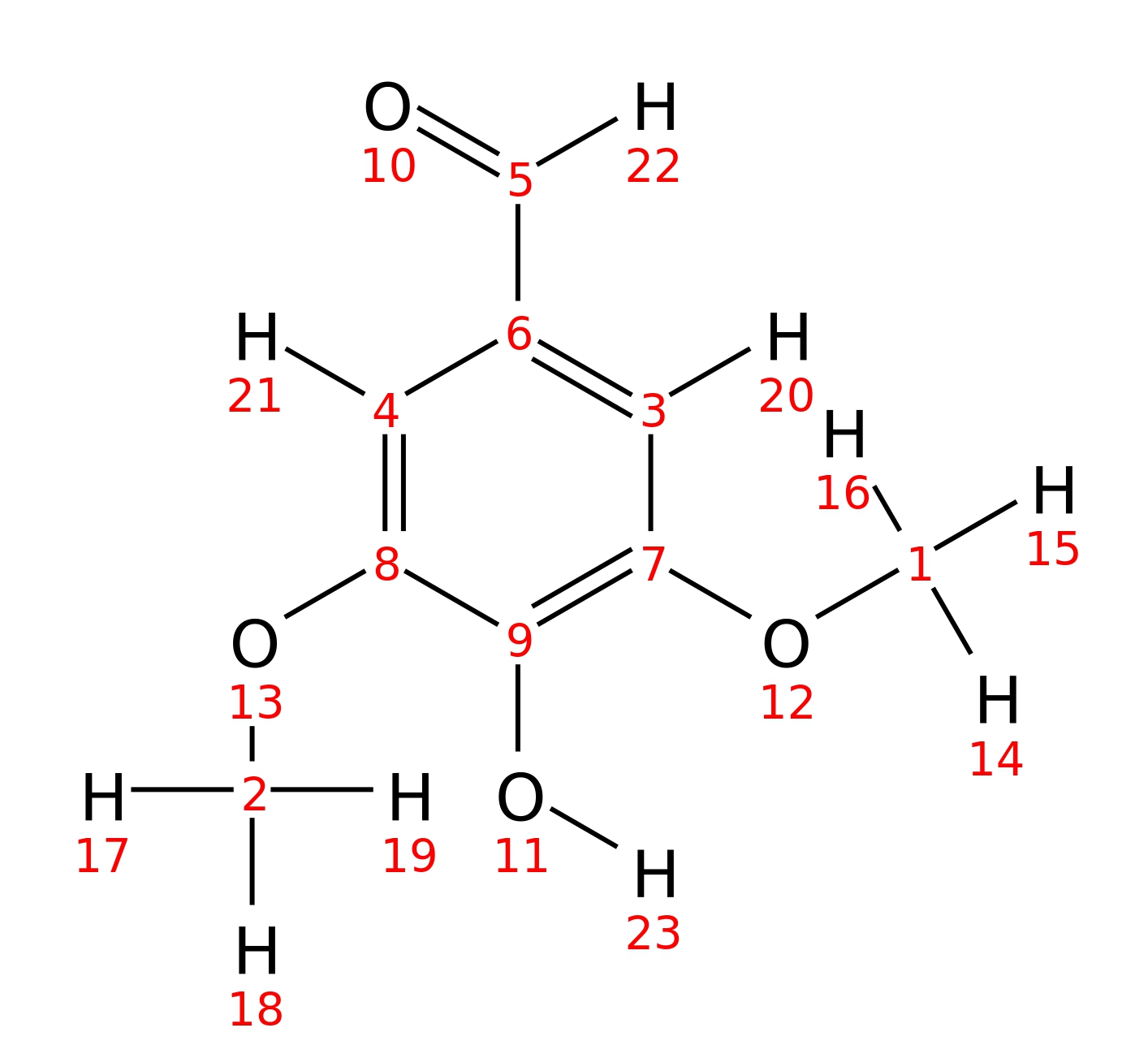
Syringaldehyde
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01318 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H10O4/c1-12-7-3-6(5-10)4-8(13-2)9(7)11/h3-5,11H,1-2H3 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| syringaldehyde | Solute | Saturated1 |
| DMSO | Solvent | 100% |
| TMS | Reference | 0.01% |
Spin System Matrix
| 14 | 15 | 16 | 20 | 21 | 17 | 18 | 19 | 22 | |
|---|---|---|---|---|---|---|---|---|---|
| 14 | 3.846 | -11.5 | -11.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 3.846 | -11.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 3.846 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 7.211 | 2.0 | 0 | 0 | 0 | 1.0 |
| 21 | 0 | 0 | 0 | 0 | 7.211 | 0 | 0 | 0 | 1.0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 3.846 | -11.5 | -11.5 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 3.846 | -11.5 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.846 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9.779 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.84584 | 1.0 | standard |
| 7.21003 | 0.198245 | standard |
| 7.21195 | 0.198112 | standard |
| 9.77746 | 0.0621164 | standard |
| 9.77917 | 0.099333 | standard |
| 9.78088 | 0.0623285 | standard |
| -0.942129 | 1.125e-06 | standard |
| 3.84584 | 1.0 | standard |
| 7.19953 | 0.200125 | standard |
| 7.22234 | 0.200017 | standard |
| 9.75746 | 0.061841 | standard |
| 9.77926 | 0.099143 | standard |
| 9.80107 | 0.06177 | standard |
| 3.84584 | 1.0 | standard |
| 7.20338 | 0.20063 | standard |
| 7.21859 | 0.20072 | standard |
| 9.76458 | 0.061707 | standard |
| 9.77922 | 0.0989913 | standard |
| 9.79368 | 0.0618782 | standard |
| 3.84584 | 1.0 | standard |
| 7.20529 | 0.200393 | standard |
| 7.21669 | 0.200498 | standard |
| 9.76824 | 0.0615311 | standard |
| 9.77923 | 0.0990226 | standard |
| 9.79006 | 0.0620141 | standard |
| 3.84584 | 1.0 | standard |
| 7.20592 | 0.200606 | standard |
| 7.21606 | 0.200409 | standard |
| 9.76948 | 0.0616716 | standard |
| 9.77917 | 0.099009 | standard |
| 9.78886 | 0.0617176 | standard |
| 3.84584 | 1.0 | standard |
| 7.20641 | 0.200396 | standard |
| 7.21556 | 0.200386 | standard |
| 9.77041 | 0.0616588 | standard |
| 9.77917 | 0.098901 | standard |
| 9.78793 | 0.0616948 | standard |
| 3.84584 | 1.0 | standard |
| 7.20867 | 0.199732 | standard |
| 7.2133 | 0.199749 | standard |
| 9.77488 | 0.0621218 | standard |
| 9.77917 | 0.099294 | standard |
| 9.78346 | 0.0620343 | standard |
| 3.84584 | 1.0 | standard |
| 7.2095 | 0.201411 | standard |
| 7.21247 | 0.201268 | standard |
| 9.7762 | 0.0612556 | standard |
| 9.77917 | 0.098702 | standard |
| 9.78214 | 0.0612865 | standard |
| 3.84584 | 1.0 | standard |
| 7.20986 | 0.201116 | standard |
| 7.21211 | 0.200745 | standard |
| 9.77703 | 0.0620514 | standard |
| 9.77917 | 0.099147 | standard |
| 9.78131 | 0.0619816 | standard |
| 3.84584 | 1.0 | standard |
| 7.21003 | 0.198404 | standard |
| 7.21195 | 0.198266 | standard |
| 9.77746 | 0.0622207 | standard |
| 9.77917 | 0.099291 | standard |
| 9.78088 | 0.0621476 | standard |
| 3.84584 | 1.0 | standard |
| 7.21022 | 0.199966 | standard |
| 7.21176 | 0.199545 | standard |
| 9.77769 | 0.0612933 | standard |
| 9.77917 | 0.098748 | standard |
| 9.78065 | 0.0614144 | standard |
| 3.84584 | 1.0 | standard |
| 7.2103 | 0.198291 | standard |
| 7.21167 | 0.19803 | standard |
| 9.77801 | 0.0631034 | standard |
| 9.77917 | 0.099967 | standard |
| 9.78032 | 0.0629745 | standard |
| 3.84584 | 1.0 | standard |
| 7.21041 | 0.202345 | standard |
| 7.21157 | 0.202109 | standard |
| 9.77793 | 0.060308 | standard |
| 9.77917 | 0.0987721 | standard |
| 9.78024 | 0.0630459 | standard |
| 3.84584 | 1.0 | standard |
| 7.21039 | 0.198711 | standard |
| 7.21158 | 0.198684 | standard |
| 9.77802 | 0.0607836 | standard |
| 9.77917 | 0.09834 | standard |
| 9.78032 | 0.0607923 | standard |
| 3.84584 | 1.0 | standard |
| 7.21049 | 0.201416 | standard |
| 7.21148 | 0.201336 | standard |
| 9.77826 | 0.0624619 | standard |
| 9.77917 | 0.09958 | standard |
| 9.78008 | 0.0624832 | standard |
| 3.84584 | 1.0 | standard |
| 7.21049 | 0.198448 | standard |
| 7.21149 | 0.198477 | standard |
| 9.7782 | 0.060511 | standard |
| 9.77917 | 0.098097 | standard |
| 9.78014 | 0.0605688 | standard |
| 3.84584 | 1.0 | standard |
| 7.21048 | 0.195764 | standard |
| 7.2115 | 0.195839 | standard |
| 9.77832 | 0.0622897 | standard |
| 9.77917 | 0.099345 | standard |
| 9.78002 | 0.0622437 | standard |
| 3.84584 | 1.0 | standard |
| 7.21058 | 0.201614 | standard |
| 7.21139 | 0.201675 | standard |
| 9.77842 | 0.0627062 | standard |
| 9.77917 | 0.099476 | standard |
| 9.77992 | 0.0626246 | standard |
| 3.84584 | 1.0 | standard |
| 7.21069 | 0.20701 | standard |
| 7.21129 | 0.206955 | standard |
| 9.77848 | 0.0606314 | standard |
| 9.77917 | 0.098256 | standard |
| 9.77986 | 0.0605692 | standard |