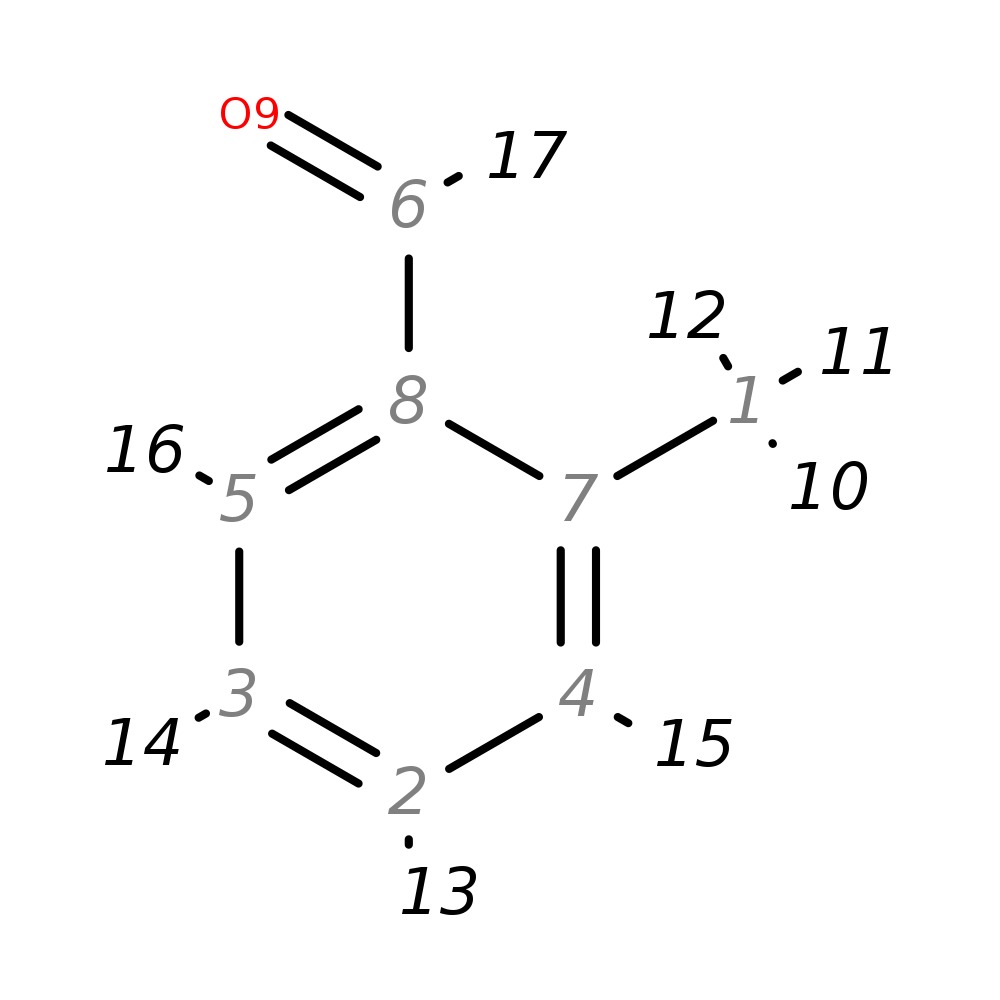
O-tolualdehyde
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.02072 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H8O/c1-7-4-2-3-5-8(7)6-9/h2-6H,1H3 | |
| Note 1 | Peak 16 has contamination of CDCl3. Oscar Li |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| o-tolualdehyde | Solute | Saturated1 |
| CDCl3 | Solvent | 100% |
| TMS | Reference | 0.5% |
Spin System Matrix
| 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|---|---|
| 10 | 2.675 | -12.4 | 0 | 0 | 0 | 0.769 | 0 | 0 |
| 11 | 0 | 2.675 | 0 | 0 | 0 | 0.769 | 0 | 0 |
| 12 | 0 | 0 | 2.675 | 0 | 0 | 0.769 | 0 | 0 |
| 13 | 0 | 0 | 0 | 7.363 | 7.101 | 8.051 | 0 | 0 |
| 14 | 0 | 0 | 0 | 0 | 7.477 | 0 | 7.825 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 7.799 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 7.263 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10.273 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.6748 | 1.00026 | standard |
| 7.25471 | 0.18643 | standard |
| 7.2703 | 0.214824 | standard |
| 7.34723 | 0.0878262 | standard |
| 7.36193 | 0.157051 | standard |
| 7.37742 | 0.118766 | standard |
| 7.46322 | 0.122888 | standard |
| 7.47817 | 0.170564 | standard |
| 7.49297 | 0.0841008 | standard |
| 7.79139 | 0.149588 | standard |
| 7.8074 | 0.139264 | standard |
| 10.273 | 0.395602 | standard |
| -0.993408 | 1.7125e-05 | standard |
| 2.67479 | 1.0 | standard |
| 7.08562 | 0.0303801 | standard |
| 7.10476 | 0.0309141 | standard |
| 7.19264 | 0.0758571 | standard |
| 7.31811 | 0.296871 | standard |
| 7.36621 | 0.245627 | standard |
| 7.4266 | 0.3822 | standard |
| 7.58413 | 0.0764073 | standard |
| 7.72586 | 0.132985 | standard |
| 7.91779 | 0.0615531 | standard |
| 10.273 | 0.391999 | standard |
| -0.996285 | 8.125e-06 | standard |
| 2.67479 | 1.0 | standard |
| 7.17002 | 0.0645614 | standard |
| 7.23368 | 0.0729841 | standard |
| 7.31291 | 0.289208 | standard |
| 7.39742 | 0.203709 | standard |
| 7.42413 | 0.330186 | standard |
| 7.4567 | 0.160506 | standard |
| 7.52975 | 0.0736401 | standard |
| 7.62693 | 0.03405 | standard |
| 7.67101 | 0.0265111 | standard |
| 7.75086 | 0.141261 | standard |
| 7.87092 | 0.083274 | standard |
| 10.273 | 0.39654 | standard |
| -0.947485 | 4.125e-06 | standard |
| 2.67481 | 1.0 | standard |
| 7.20114 | 0.0902043 | standard |
| 7.26425 | 0.0809121 | standard |
| 7.30047 | 0.22322 | standard |
| 7.33672 | 0.149975 | standard |
| 7.42159 | 0.302299 | standard |
| 7.5067 | 0.099569 | standard |
| 7.58774 | 0.035025 | standard |
| 7.6848 | 0.0132911 | standard |
| 7.75957 | 0.151803 | standard |
| 7.85071 | 0.100394 | standard |
| 10.273 | 0.398083 | standard |
| -0.922193 | 3.125e-06 | standard |
| 2.67479 | 1.0 | standard |
| 7.21022 | 0.101611 | standard |
| 7.29663 | 0.223588 | standard |
| 7.34208 | 0.145691 | standard |
| 7.41797 | 0.300363 | standard |
| 7.42579 | 0.288442 | standard |
| 7.5008 | 0.111447 | standard |
| 7.54773 | 0.0148231 | standard |
| 7.57478 | 0.0377071 | standard |
| 7.68956 | 0.0103941 | standard |
| 7.76312 | 0.154866 | standard |
| 7.84452 | 0.106904 | standard |
| 10.273 | 0.399009 | standard |
| -0.974761 | 3.125e-06 | standard |
| 2.6748 | 1.00001 | standard |
| 7.21694 | 0.111609 | standard |
| 7.29318 | 0.232887 | standard |
| 7.3459 | 0.145991 | standard |
| 7.42285 | 0.288824 | standard |
| 7.49655 | 0.122104 | standard |
| 7.56443 | 0.04039 | standard |
| 7.69293 | 0.00810212 | standard |
| 7.76605 | 0.156703 | standard |
| 7.83972 | 0.112474 | standard |
| 10.273 | 0.398828 | standard |
| -0.917332 | 1.125e-06 | standard |
| 2.6748 | 1.00004 | standard |
| 7.24188 | 0.164272 | standard |
| 7.28009 | 0.232582 | standard |
| 7.32214 | 0.0728075 | standard |
| 7.35805 | 0.15786 | standard |
| 7.39639 | 0.146667 | standard |
| 7.44526 | 0.158852 | standard |
| 7.48234 | 0.16495 | standard |
| 7.51852 | 0.064223 | standard |
| 7.78057 | 0.158656 | standard |
| 7.81978 | 0.133463 | standard |
| 10.273 | 0.400643 | standard |
| 2.6748 | 1.00009 | standard |
| 7.24911 | 0.175863 | standard |
| 7.27484 | 0.223595 | standard |
| 7.33628 | 0.0789672 | standard |
| 7.36048 | 0.155881 | standard |
| 7.38633 | 0.1312 | standard |
| 7.4547 | 0.138571 | standard |
| 7.47961 | 0.167307 | standard |
| 7.50407 | 0.0740946 | standard |
| 7.78642 | 0.152173 | standard |
| 7.8129 | 0.135804 | standard |
| 10.273 | 0.395594 | standard |
| 2.6748 | 1.00016 | standard |
| 7.2526 | 0.183731 | standard |
| 7.27206 | 0.219467 | standard |
| 7.34316 | 0.0850507 | standard |
| 7.36144 | 0.156789 | standard |
| 7.38084 | 0.123876 | standard |
| 7.4599 | 0.12905 | standard |
| 7.47864 | 0.170052 | standard |
| 7.49703 | 0.0805978 | standard |
| 7.78952 | 0.151225 | standard |
| 7.80948 | 0.139031 | standard |
| 10.273 | 0.397272 | standard |
| 2.6748 | 1.00026 | standard |
| 7.25471 | 0.186451 | standard |
| 7.2703 | 0.214839 | standard |
| 7.34723 | 0.0878372 | standard |
| 7.36193 | 0.157047 | standard |
| 7.37742 | 0.11877 | standard |
| 7.46322 | 0.122891 | standard |
| 7.47817 | 0.170566 | standard |
| 7.49297 | 0.0841129 | standard |
| 7.79139 | 0.149579 | standard |
| 7.8074 | 0.139259 | standard |
| 10.273 | 0.395642 | standard |
| 2.6748 | 1.00033 | standard |
| 7.25607 | 0.191215 | standard |
| 7.26906 | 0.215247 | standard |
| 7.3499 | 0.0918151 | standard |
| 7.36221 | 0.158907 | standard |
| 7.37514 | 0.116349 | standard |
| 7.46545 | 0.120768 | standard |
| 7.47791 | 0.172129 | standard |
| 7.49029 | 0.0878839 | standard |
| 7.79268 | 0.147848 | standard |
| 7.80604 | 0.142593 | standard |
| 10.273 | 0.400801 | standard |
| 2.6748 | 1.00048 | standard |
| 7.25706 | 0.192021 | standard |
| 7.26817 | 0.212539 | standard |
| 7.35184 | 0.0933954 | standard |
| 7.36234 | 0.157383 | standard |
| 7.3734 | 0.113293 | standard |
| 7.46709 | 0.117395 | standard |
| 7.47779 | 0.173137 | standard |
| 7.48836 | 0.0896463 | standard |
| 7.79362 | 0.14661 | standard |
| 7.80504 | 0.143876 | standard |
| 10.273 | 0.399116 | standard |
| 2.6748 | 1.00042 | standard |
| 7.25746 | 0.197687 | standard |
| 7.26787 | 0.216529 | standard |
| 7.35258 | 0.0967621 | standard |
| 7.36243 | 0.161327 | standard |
| 7.37276 | 0.115027 | standard |
| 7.46773 | 0.119359 | standard |
| 7.47772 | 0.176223 | standard |
| 7.48761 | 0.092845 | standard |
| 7.79396 | 0.150684 | standard |
| 7.80465 | 0.149097 | standard |
| 10.273 | 0.408787 | standard |
| 2.6748 | 1.00076 | standard |
| 7.25776 | 0.189949 | standard |
| 7.26757 | 0.206079 | standard |
| 7.35327 | 0.0930351 | standard |
| 7.36244 | 0.153296 | standard |
| 7.37216 | 0.108958 | standard |
| 7.46833 | 0.112878 | standard |
| 7.47769 | 0.168408 | standard |
| 7.48697 | 0.0896512 | standard |
| 7.79428 | 0.143207 | standard |
| 7.80434 | 0.142426 | standard |
| 10.273 | 0.390968 | standard |
| 2.6748 | 1.00066 | standard |
| 7.25835 | 0.198241 | standard |
| 7.26703 | 0.212559 | standard |
| 7.35436 | 0.0974691 | standard |
| 7.3625 | 0.157143 | standard |
| 7.37112 | 0.11241 | standard |
| 7.46932 | 0.115827 | standard |
| 7.47758 | 0.175285 | standard |
| 7.48588 | 0.0943132 | standard |
| 7.79483 | 0.148235 | standard |
| 7.80377 | 0.149983 | standard |
| 10.273 | 0.406157 | standard |
| 2.6748 | 1.00057 | standard |
| 7.25855 | 0.202638 | standard |
| 7.26678 | 0.217037 | standard |
| 7.35476 | 0.100214 | standard |
| 7.3626 | 0.16306 | standard |
| 7.37072 | 0.11372 | standard |
| 7.46972 | 0.117516 | standard |
| 7.47758 | 0.176011 | standard |
| 7.48538 | 0.0967853 | standard |
| 7.79505 | 0.151819 | standard |
| 7.80351 | 0.145507 | standard |
| 10.273 | 0.414396 | standard |
| 2.6748 | 1.00129 | standard |
| 7.25877 | 0.189593 | standard |
| 7.26658 | 0.202872 | standard |
| 7.35526 | 0.0941851 | standard |
| 7.36256 | 0.149514 | standard |
| 7.37033 | 0.106137 | standard |
| 7.47011 | 0.109513 | standard |
| 7.47759 | 0.162107 | standard |
| 7.48499 | 0.0910732 | standard |
| 7.79523 | 0.139896 | standard |
| 7.8033 | 0.142419 | standard |
| 10.273 | 0.38801 | standard |
| 2.6748 | 1.00121 | standard |
| 7.25914 | 0.197813 | standard |
| 7.26628 | 0.206534 | standard |
| 7.35595 | 0.0974261 | standard |
| 7.36268 | 0.158005 | standard |
| 7.36973 | 0.108578 | standard |
| 7.47071 | 0.110915 | standard |
| 7.47753 | 0.169179 | standard |
| 7.48429 | 0.0953092 | standard |
| 7.7956 | 0.145488 | standard |
| 7.80292 | 0.145489 | standard |
| 10.273 | 0.399426 | standard |
| 2.6748 | 1.00255 | standard |
| 7.25974 | 0.189992 | standard |
| 7.26574 | 0.194618 | standard |
| 7.35704 | 0.094063 | standard |
| 7.36276 | 0.15201 | standard |
| 7.36864 | 0.102537 | standard |
| 7.4717 | 0.10439 | standard |
| 7.47748 | 0.160123 | standard |
| 7.4832 | 0.0929791 | standard |
| 7.79614 | 0.138298 | standard |
| 7.80238 | 0.137963 | standard |
| 10.273 | 0.381048 | standard |