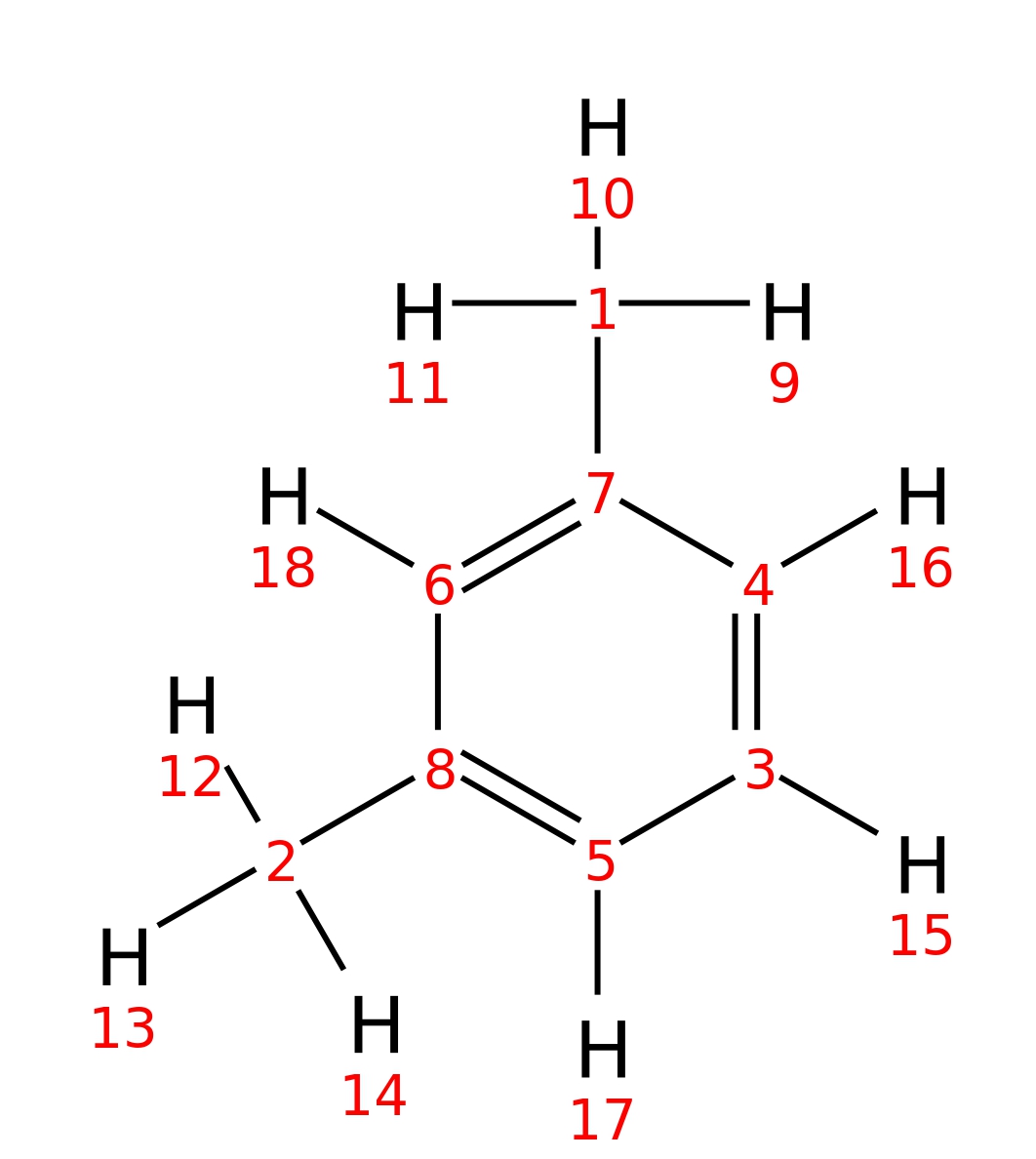
M-xylene
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01714 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H10/c1-7-4-3-5-8(2)6-7/h3-6H,1-2H3 | |
| Note 1 | 16?17 | |
| Note 2 | lineshapes -aromatic |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| m-xylene | Solute | Saturated1 |
| CDCl3 | Solvent | 100% |
| TMS | Reference | 0.5% |
Spin System Matrix
| 9 | 10 | 11 | 18 | 16 | 15 | 17 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 9 | 2.312 | -14.9 | -14.9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 2.312 | -14.9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 2.312 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 6.996 | 1.659 | 0 | 1.659 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 6.972 | 7.499 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 7.141 | 7.499 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 6.972 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.312 | -14.9 | -14.9 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.312 | -14.9 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.312 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.31222 | 1.0 | standard |
| 6.96441 | 0.0788666 | standard |
| 6.97931 | 0.0947287 | standard |
| 6.99584 | 0.0786788 | standard |
| 7.1271 | 0.031584 | standard |
| 7.14191 | 0.0490812 | standard |
| 7.15673 | 0.0232756 | standard |
| 2.31222 | 1.0 | standard |
| 7.01314 | 0.286868 | standard |
| 2.31222 | 1.0 | standard |
| 7.00547 | 0.245985 | standard |
| 7.13303 | 0.0461881 | standard |
| 7.27863 | 0.0112091 | standard |
| 2.31222 | 1.0 | standard |
| 7.00131 | 0.23001 | standard |
| 7.0699 | 0.07339 | standard |
| 7.13987 | 0.0392591 | standard |
| 7.24699 | 0.0126231 | standard |
| 2.31222 | 1.0 | standard |
| 6.99979 | 0.225915 | standard |
| 7.07644 | 0.062782 | standard |
| 7.14096 | 0.0381291 | standard |
| 7.23469 | 0.0132641 | standard |
| 2.31222 | 1.0 | standard |
| 6.94923 | 0.0605931 | standard |
| 6.99846 | 0.222996 | standard |
| 7.0815 | 0.055736 | standard |
| 7.14166 | 0.0376981 | standard |
| 7.22472 | 0.013889 | standard |
| 2.31222 | 1.0 | standard |
| 6.95292 | 0.0724104 | standard |
| 6.99097 | 0.185707 | standard |
| 7.1095 | 0.0356863 | standard |
| 7.14295 | 0.0408736 | standard |
| 7.18133 | 0.0185075 | standard |
| 2.31222 | 1.0 | standard |
| 6.95946 | 0.0764792 | standard |
| 6.98554 | 0.12471 | standard |
| 7.11883 | 0.0340969 | standard |
| 7.14276 | 0.0456121 | standard |
| 7.16753 | 0.0209928 | standard |
| 2.31222 | 1.0 | standard |
| 6.96263 | 0.0780273 | standard |
| 6.98132 | 0.102293 | standard |
| 6.99517 | 0.0837311 | standard |
| 7.12389 | 0.0328849 | standard |
| 7.14225 | 0.0477576 | standard |
| 7.16076 | 0.0222852 | standard |
| 2.31222 | 1.0 | standard |
| 6.96441 | 0.0788574 | standard |
| 6.97931 | 0.0947119 | standard |
| 6.99584 | 0.0786669 | standard |
| 7.1271 | 0.031581 | standard |
| 7.14191 | 0.0490792 | standard |
| 7.15673 | 0.0232746 | standard |
| 2.31222 | 1.0 | standard |
| 6.96561 | 0.0795106 | standard |
| 6.97793 | 0.0915998 | standard |
| 6.99596 | 0.0770786 | standard |
| 7.12937 | 0.0307621 | standard |
| 7.14175 | 0.04954 | standard |
| 7.15407 | 0.0238317 | standard |
| 2.31222 | 1.0 | standard |
| 6.9665 | 0.0801024 | standard |
| 6.977 | 0.0899619 | standard |
| 6.99601 | 0.0756578 | standard |
| 7.131 | 0.030204 | standard |
| 7.14163 | 0.0499117 | standard |
| 7.15225 | 0.0242184 | standard |
| 2.31222 | 1.0 | standard |
| 6.96682 | 0.0801441 | standard |
| 6.97662 | 0.0891629 | standard |
| 6.99605 | 0.0753813 | standard |
| 7.13164 | 0.0299796 | standard |
| 7.14158 | 0.050071 | standard |
| 7.15146 | 0.0244013 | standard |
| 2.31222 | 1.0 | standard |
| 6.96711 | 0.0804599 | standard |
| 6.97631 | 0.0888149 | standard |
| 6.99604 | 0.0753726 | standard |
| 7.13223 | 0.0297797 | standard |
| 7.14155 | 0.0501325 | standard |
| 7.15082 | 0.0245384 | standard |
| 2.31222 | 1.0 | standard |
| 6.96762 | 0.080347 | standard |
| 6.97575 | 0.0874489 | standard |
| 6.99605 | 0.0749371 | standard |
| 7.13322 | 0.029452 | standard |
| 7.14151 | 0.0502731 | standard |
| 7.14974 | 0.02479 | standard |
| 2.31222 | 1.0 | standard |
| 6.96784 | 0.0804401 | standard |
| 6.97555 | 0.0880263 | standard |
| 6.99608 | 0.0743866 | standard |
| 7.13361 | 0.0293039 | standard |
| 7.14148 | 0.0503291 | standard |
| 7.1493 | 0.0249023 | standard |
| 2.31222 | 1.0 | standard |
| 6.96799 | 0.0806642 | standard |
| 6.97529 | 0.0870201 | standard |
| 6.99609 | 0.0744167 | standard |
| 7.13401 | 0.0291973 | standard |
| 7.14146 | 0.0504368 | standard |
| 7.1489 | 0.0249868 | standard |
| 2.31222 | 1.0 | standard |
| 6.96827 | 0.0825215 | standard |
| 6.97497 | 0.087542 | standard |
| 6.99606 | 0.0747977 | standard |
| 7.13465 | 0.0289514 | standard |
| 7.14143 | 0.0504737 | standard |
| 7.14821 | 0.0251477 | standard |
| 2.31222 | 1.0 | standard |
| 6.96883 | 0.0813889 | standard |
| 6.97444 | 0.086199 | standard |
| 6.99609 | 0.074353 | standard |
| 7.13569 | 0.0286715 | standard |
| 7.14141 | 0.050556 | standard |
| 7.14713 | 0.0254297 | standard |