3-Methylbenzyl-alcohol
Simulation outputs:
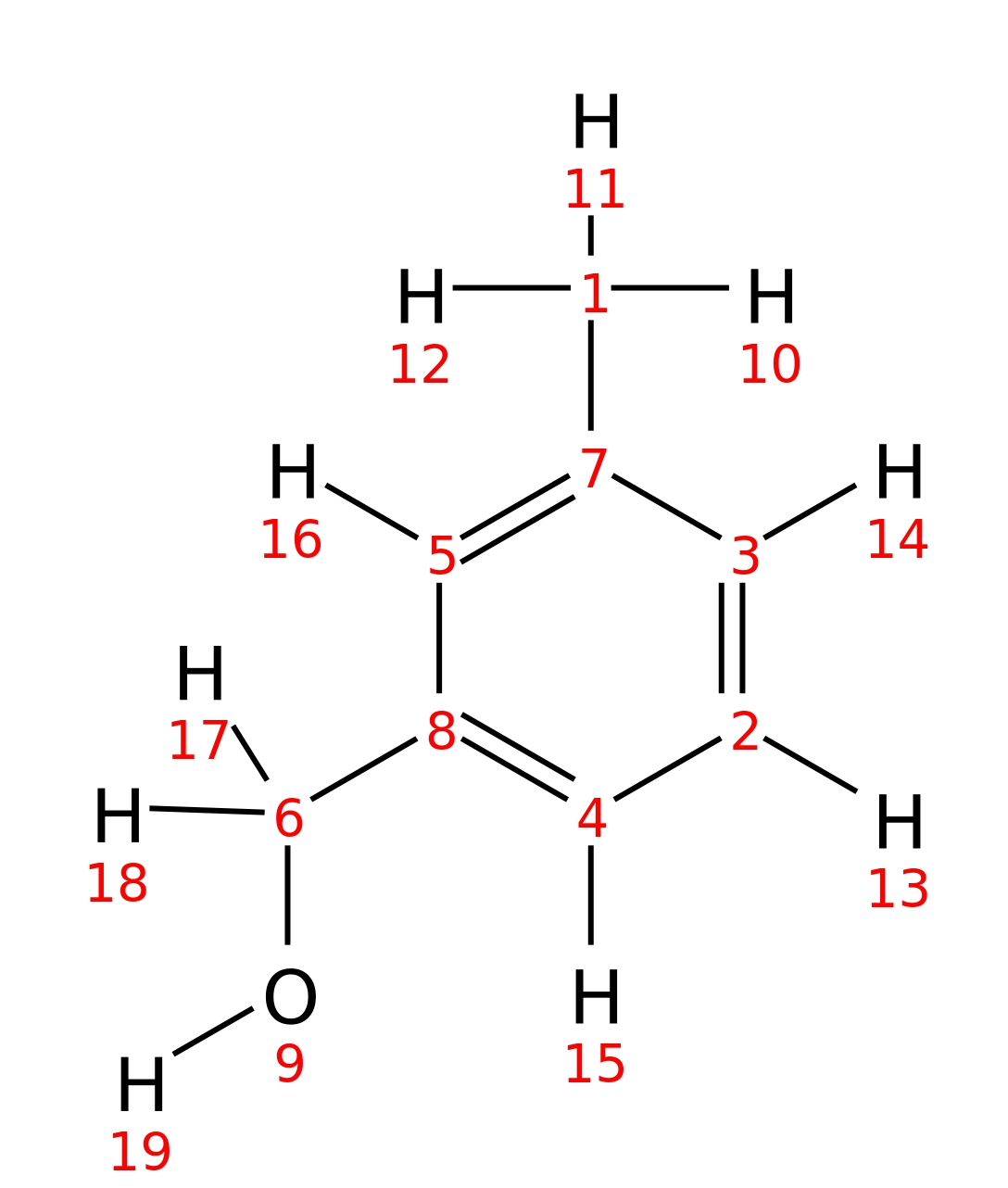
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.03040 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H10O/c1-7-3-2-4-8(5-7)6-9/h2-5,9H,6H2,1H3 | |
| Note 1 | 17,18 saturated |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-methylbenzyl alcohol | Solute | Saturated1 |
| CDCl3 | Solvent | 100% |
| TMS | Reference | 0.5% |
Spin System Matrix
| 10 | 11 | 12 | 16 | 14 | 13 | 15 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|
| 10 | 2.355 | -14.9 | -14.9 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 2.355 | -14.9 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 2.355 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 7.179 | 2.106 | 0 | 1.472 | 0 | 0 |
| 14 | 0 | 0 | 0 | 0 | 7.148 | 8.022 | 1.998 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 7.246 | 7.256 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 7.106 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.782 | -12.4 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.782 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.665197 | standard |
| 7.09847 | 0.0844274 | standard |
| 7.11267 | 0.102041 | standard |
| 7.13986 | 0.080069 | standard |
| 7.1558 | 0.109458 | standard |
| 7.17853 | 0.178053 | standard |
| 7.23223 | 0.0943543 | standard |
| 7.24724 | 0.126113 | standard |
| 7.26254 | 0.0573425 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.666414 | standard |
| 7.16925 | 0.905397 | standard |
| 7.49833 | 0.0140071 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.665968 | standard |
| 7.17083 | 0.72377 | standard |
| 7.41015 | 0.0177121 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.666688 | standard |
| 7.17178 | 0.557512 | standard |
| 7.36661 | 0.0212281 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.665802 | standard |
| 7.17159 | 0.492639 | standard |
| 7.25597 | 0.076712 | standard |
| 7.35213 | 0.0224962 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.665245 | standard |
| 7.17122 | 0.441984 | standard |
| 7.25473 | 0.0770168 | standard |
| 7.34043 | 0.0240173 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.666672 | standard |
| 7.08751 | 0.0723556 | standard |
| 7.0942 | 0.069446 | standard |
| 7.12386 | 0.163044 | standard |
| 7.17474 | 0.263794 | standard |
| 7.21583 | 0.131164 | standard |
| 7.25105 | 0.103111 | standard |
| 7.28915 | 0.0393977 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.664619 | standard |
| 7.09338 | 0.0785189 | standard |
| 7.11724 | 0.112328 | standard |
| 7.12138 | 0.107513 | standard |
| 7.13307 | 0.0832171 | standard |
| 7.13905 | 0.0615721 | standard |
| 7.15989 | 0.13285 | standard |
| 7.17812 | 0.190493 | standard |
| 7.22423 | 0.11039 | standard |
| 7.24871 | 0.11736 | standard |
| 7.27412 | 0.048484 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.665426 | standard |
| 7.09646 | 0.082331 | standard |
| 7.11426 | 0.104186 | standard |
| 7.13749 | 0.0789774 | standard |
| 7.1418 | 0.058975 | standard |
| 7.15741 | 0.116721 | standard |
| 7.17845 | 0.181367 | standard |
| 7.22905 | 0.0999273 | standard |
| 7.24775 | 0.123509 | standard |
| 7.26674 | 0.0539848 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.666668 | standard |
| 7.09847 | 0.0844904 | standard |
| 7.11267 | 0.102112 | standard |
| 7.13986 | 0.0801319 | standard |
| 7.1558 | 0.109536 | standard |
| 7.17853 | 0.178183 | standard |
| 7.23223 | 0.0944293 | standard |
| 7.24724 | 0.126223 | standard |
| 7.26249 | 0.0573491 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.665888 | standard |
| 7.09967 | 0.0852923 | standard |
| 7.11166 | 0.100213 | standard |
| 7.1414 | 0.0814278 | standard |
| 7.1547 | 0.105224 | standard |
| 7.17852 | 0.175896 | standard |
| 7.23441 | 0.091164 | standard |
| 7.24695 | 0.12775 | standard |
| 7.25967 | 0.060152 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.665489 | standard |
| 7.10061 | 0.0856264 | standard |
| 7.11079 | 0.0990056 | standard |
| 7.14248 | 0.0825592 | standard |
| 7.15386 | 0.10256 | standard |
| 7.17852 | 0.175974 | standard |
| 7.23599 | 0.0884306 | standard |
| 7.24679 | 0.127144 | standard |
| 7.25769 | 0.0618621 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.666666 | standard |
| 7.10097 | 0.0876091 | standard |
| 7.11049 | 0.0984571 | standard |
| 7.14288 | 0.0830711 | standard |
| 7.15357 | 0.102144 | standard |
| 7.17853 | 0.174637 | standard |
| 7.23664 | 0.0874924 | standard |
| 7.24675 | 0.127827 | standard |
| 7.25695 | 0.0626492 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.666103 | standard |
| 7.10125 | 0.0874333 | standard |
| 7.11023 | 0.0985656 | standard |
| 7.14328 | 0.0833472 | standard |
| 7.15327 | 0.10106 | standard |
| 7.17855 | 0.175191 | standard |
| 7.23723 | 0.0865565 | standard |
| 7.24671 | 0.128689 | standard |
| 7.25625 | 0.0632711 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.666343 | standard |
| 7.1018 | 0.0876952 | standard |
| 7.10981 | 0.0983821 | standard |
| 7.14386 | 0.0841204 | standard |
| 7.15278 | 0.0998883 | standard |
| 7.17859 | 0.175046 | standard |
| 7.23817 | 0.0849584 | standard |
| 7.2466 | 0.129913 | standard |
| 7.25506 | 0.064367 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.666361 | standard |
| 7.10198 | 0.0888014 | standard |
| 7.10961 | 0.0974601 | standard |
| 7.14417 | 0.0840582 | standard |
| 7.15258 | 0.0988248 | standard |
| 7.1786 | 0.174262 | standard |
| 7.23862 | 0.0842637 | standard |
| 7.2466 | 0.128363 | standard |
| 7.25462 | 0.064584 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.66582 | standard |
| 7.10223 | 0.0892043 | standard |
| 7.10938 | 0.097534 | standard |
| 7.14436 | 0.0847549 | standard |
| 7.15237 | 0.0989261 | standard |
| 7.17855 | 0.175759 | standard |
| 7.23897 | 0.0836506 | standard |
| 7.24652 | 0.128838 | standard |
| 7.25417 | 0.0651329 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.666094 | standard |
| 7.10251 | 0.0888017 | standard |
| 7.10915 | 0.0975761 | standard |
| 7.14475 | 0.0854972 | standard |
| 7.15202 | 0.0976965 | standard |
| 7.17853 | 0.174675 | standard |
| 7.23961 | 0.0828518 | standard |
| 7.2465 | 0.128098 | standard |
| 7.25348 | 0.0658707 | standard |
| 2.35537 | 1.0 | standard |
| 4.78167 | 0.666171 | standard |
| 7.10304 | 0.0899274 | standard |
| 7.10857 | 0.0968306 | standard |
| 7.14534 | 0.0866008 | standard |
| 7.15147 | 0.0965556 | standard |
| 7.1786 | 0.175001 | standard |
| 7.2406 | 0.0815019 | standard |
| 7.24643 | 0.129879 | standard |
| 7.25229 | 0.0671539 | standard |