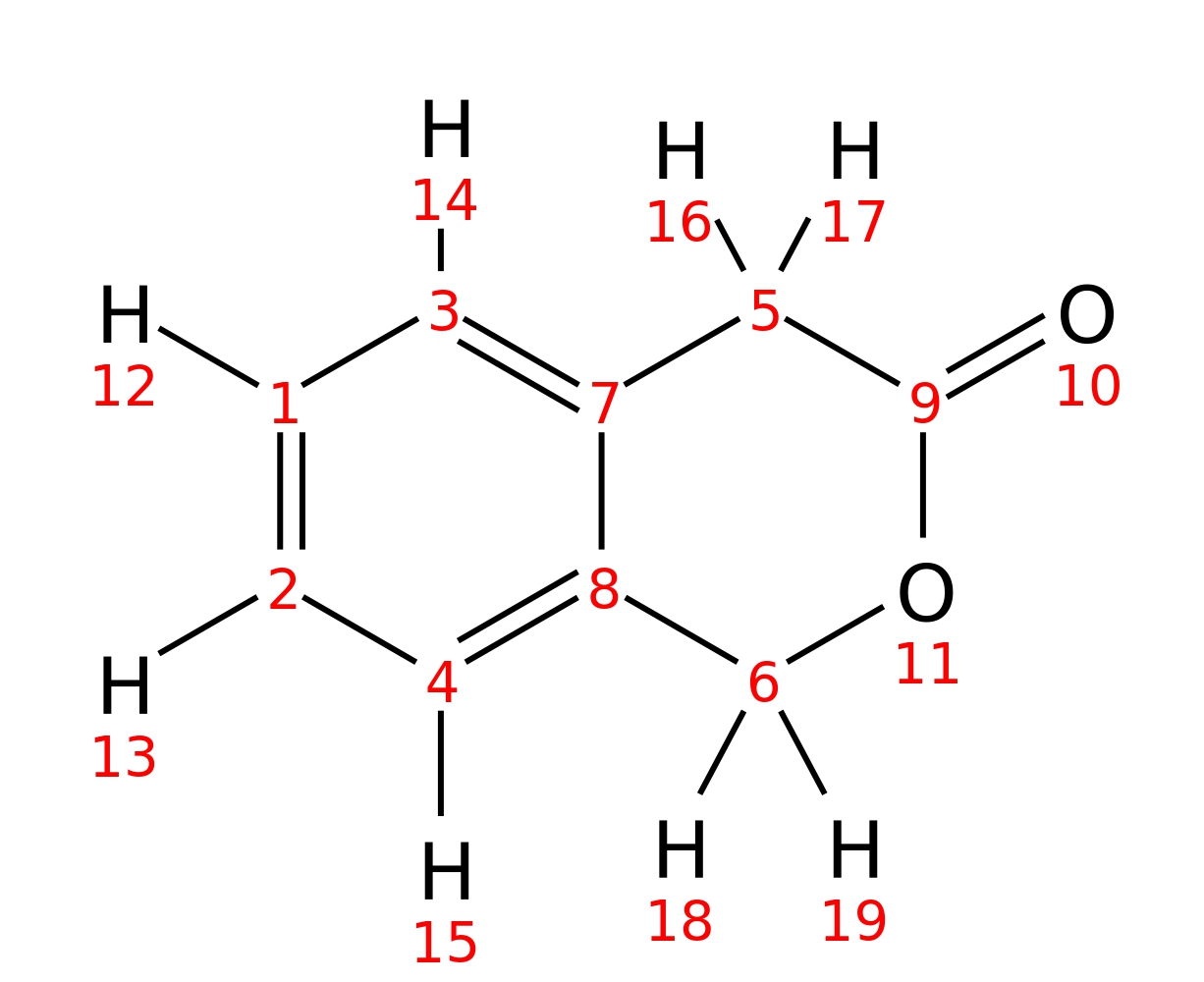
3-Isochromanone
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.01285 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H8O2/c10-9-5-7-3-1-2-4-8(7)6-11-9/h1-4H,5-6H2 | |
| Note 1 | 16,17?18,19 | |
| Note 2 | 14?15 | |
| Note 3 | 12?13 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-isochromanone | Solute | Saturated1 |
| CDCl3 | Solvent | 100% |
| TMS | Reference | 0.5% |
Spin System Matrix
| 16 | 17 | 14 | 12 | 13 | 15 | 18 | 19 | |
|---|---|---|---|---|---|---|---|---|
| 16 | 3.718 | -16.16 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 3.718 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 7.252 | 7.528 | 1.078 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 7.347 | 7.491 | 1.078 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 7.312 | 7.491 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 7.228 | 1.0 | 1.0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 5.319 | -15.63 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.319 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.7175 | 1.0 | standard |
| 5.3187 | 0.805619 | standard |
| 7.21876 | 0.132029 | standard |
| 7.2396 | 0.256265 | standard |
| 7.25825 | 0.193779 | standard |
| 7.26142 | 0.208619 | standard |
| 7.29374 | 0.0804033 | standard |
| 7.30934 | 0.179296 | standard |
| 7.31175 | 0.173163 | standard |
| 7.33038 | 0.247419 | standard |
| 7.34868 | 0.155096 | standard |
| 7.36692 | 0.0497816 | standard |
| -0.912472 | 1.125e-06 | standard |
| 3.7175 | 0.737281 | standard |
| 5.31843 | 0.67783 | standard |
| 7.06885 | 0.0307121 | standard |
| 7.2867 | 1.0 | standard |
| 7.50081 | 0.0292641 | standard |
| -0.977439 | 1.125e-06 | standard |
| 3.7175 | 0.973675 | standard |
| 5.31852 | 0.882344 | standard |
| 7.14106 | 0.0600201 | standard |
| 7.28879 | 1.0 | standard |
| 7.43862 | 0.0534101 | standard |
| 3.7175 | 1.0 | standard |
| 5.31864 | 0.887675 | standard |
| 7.18728 | 0.0994702 | standard |
| 7.29338 | 0.77908 | standard |
| 7.40604 | 0.07261 | standard |
| 3.7175 | 1.0 | standard |
| 5.31867 | 0.877533 | standard |
| 7.19459 | 0.116947 | standard |
| 7.28092 | 0.623293 | standard |
| 7.29479 | 0.67478 | standard |
| 7.39495 | 0.0806651 | standard |
| 3.7175 | 1.0 | standard |
| 5.31871 | 0.868566 | standard |
| 7.20114 | 0.130822 | standard |
| 7.27873 | 0.551131 | standard |
| 7.29631 | 0.585496 | standard |
| 7.34055 | 0.20751 | standard |
| 7.38581 | 0.08914 | standard |
| 3.7175 | 1.0 | standard |
| 5.31869 | 0.817952 | standard |
| 7.2434 | 0.273095 | standard |
| 7.26944 | 0.293688 | standard |
| 7.30444 | 0.245016 | standard |
| 7.31837 | 0.261248 | standard |
| 7.3333 | 0.275195 | standard |
| 7.35151 | 0.156512 | standard |
| 7.38837 | 0.0315742 | standard |
| 3.7175 | 1.0 | standard |
| 5.31866 | 0.805665 | standard |
| 7.2157 | 0.118962 | standard |
| 7.23682 | 0.295485 | standard |
| 7.24017 | 0.287644 | standard |
| 7.25891 | 0.185109 | standard |
| 7.26429 | 0.219967 | standard |
| 7.28799 | 0.0874385 | standard |
| 7.30783 | 0.189348 | standard |
| 7.3121 | 0.172849 | standard |
| 7.32888 | 0.2544 | standard |
| 7.35006 | 0.144234 | standard |
| 7.37406 | 0.0417619 | standard |
| 3.7175 | 1.0 | standard |
| 5.3187 | 0.805721 | standard |
| 7.21868 | 0.132116 | standard |
| 7.23959 | 0.256428 | standard |
| 7.25825 | 0.193821 | standard |
| 7.26142 | 0.208662 | standard |
| 7.29368 | 0.0805171 | standard |
| 7.30934 | 0.179319 | standard |
| 7.31174 | 0.173186 | standard |
| 7.33039 | 0.247353 | standard |
| 7.34868 | 0.155112 | standard |
| 7.36692 | 0.0497816 | standard |
| 3.7175 | 1.0 | standard |
| 5.31866 | 0.8026 | standard |
| 7.22062 | 0.139994 | standard |
| 7.2352 | 0.200341 | standard |
| 7.24299 | 0.170281 | standard |
| 7.24506 | 0.166675 | standard |
| 7.25769 | 0.200802 | standard |
| 7.25954 | 0.20847 | standard |
| 7.29718 | 0.0817653 | standard |
| 7.31015 | 0.179922 | standard |
| 7.31168 | 0.176579 | standard |
| 7.32431 | 0.129732 | standard |
| 7.32655 | 0.121234 | standard |
| 7.3333 | 0.15768 | standard |
| 7.33537 | 0.155672 | standard |
| 7.34806 | 0.1645 | standard |
| 7.36268 | 0.0559267 | standard |
| 3.7175 | 1.0 | standard |
| 5.31871 | 0.796258 | standard |
| 7.22176 | 0.145819 | standard |
| 7.23388 | 0.193314 | standard |
| 7.24479 | 0.166546 | standard |
| 7.24613 | 0.168273 | standard |
| 7.25708 | 0.203285 | standard |
| 7.25823 | 0.207442 | standard |
| 7.29958 | 0.0827838 | standard |
| 7.31062 | 0.183189 | standard |
| 7.31154 | 0.181119 | standard |
| 7.32249 | 0.119105 | standard |
| 7.33544 | 0.145226 | standard |
| 7.33666 | 0.146467 | standard |
| 7.34766 | 0.169416 | standard |
| 7.36005 | 0.0621976 | standard |
| 3.7175 | 1.0 | standard |
| 5.31866 | 0.808929 | standard |
| 7.22285 | 0.146706 | standard |
| 7.23321 | 0.190703 | standard |
| 7.24596 | 0.168746 | standard |
| 7.2469 | 0.170581 | standard |
| 7.2566 | 0.204565 | standard |
| 7.25732 | 0.206664 | standard |
| 7.30126 | 0.0848489 | standard |
| 7.311 | 0.187624 | standard |
| 7.32121 | 0.115806 | standard |
| 7.33778 | 0.139754 | standard |
| 7.34746 | 0.174501 | standard |
| 7.35805 | 0.0661736 | standard |
| 3.7175 | 1.0 | standard |
| 5.31869 | 0.798559 | standard |
| 7.22308 | 0.149754 | standard |
| 7.23288 | 0.188645 | standard |
| 7.24716 | 0.173991 | standard |
| 7.25693 | 0.206815 | standard |
| 7.30197 | 0.0848815 | standard |
| 7.31105 | 0.18676 | standard |
| 7.32071 | 0.115383 | standard |
| 7.33754 | 0.134174 | standard |
| 7.33831 | 0.135716 | standard |
| 7.34746 | 0.179569 | standard |
| 7.35723 | 0.0676751 | standard |
| 3.7175 | 1.0 | standard |
| 5.31868 | 0.802504 | standard |
| 7.22344 | 0.15168 | standard |
| 7.23257 | 0.187089 | standard |
| 7.2474 | 0.176633 | standard |
| 7.25659 | 0.207809 | standard |
| 7.30253 | 0.0872423 | standard |
| 7.31114 | 0.187034 | standard |
| 7.32028 | 0.115743 | standard |
| 7.33866 | 0.136332 | standard |
| 7.34735 | 0.180466 | standard |
| 7.35651 | 0.0692 | standard |
| 3.7175 | 1.0 | standard |
| 5.31871 | 0.805784 | standard |
| 7.22406 | 0.152964 | standard |
| 7.23202 | 0.183574 | standard |
| 7.24789 | 0.17627 | standard |
| 7.25557 | 0.200327 | standard |
| 7.25619 | 0.200608 | standard |
| 7.30353 | 0.0889378 | standard |
| 7.31123 | 0.186724 | standard |
| 7.31158 | 0.186366 | standard |
| 7.31933 | 0.111066 | standard |
| 7.33955 | 0.128671 | standard |
| 7.3473 | 0.184277 | standard |
| 7.35542 | 0.0726056 | standard |
| 3.7175 | 1.0 | standard |
| 5.31869 | 0.799498 | standard |
| 7.22427 | 0.15386 | standard |
| 7.23196 | 0.186838 | standard |
| 7.24808 | 0.178396 | standard |
| 7.25572 | 0.209345 | standard |
| 7.30391 | 0.0901674 | standard |
| 7.31138 | 0.189853 | standard |
| 7.31909 | 0.114424 | standard |
| 7.33985 | 0.127817 | standard |
| 7.34722 | 0.183155 | standard |
| 7.35503 | 0.0744677 | standard |
| 3.7175 | 1.0 | standard |
| 5.31866 | 0.802606 | standard |
| 7.22444 | 0.157262 | standard |
| 7.23176 | 0.185003 | standard |
| 7.24827 | 0.178051 | standard |
| 7.25532 | 0.203416 | standard |
| 7.25564 | 0.203534 | standard |
| 7.30436 | 0.0886561 | standard |
| 7.31152 | 0.192919 | standard |
| 7.31865 | 0.111326 | standard |
| 7.34013 | 0.127688 | standard |
| 7.34713 | 0.183415 | standard |
| 7.35466 | 0.0766331 | standard |
| 3.7175 | 1.0 | standard |
| 5.31871 | 0.8058 | standard |
| 7.22476 | 0.158568 | standard |
| 7.23136 | 0.180263 | standard |
| 7.2485 | 0.185912 | standard |
| 7.25514 | 0.21183 | standard |
| 7.30492 | 0.0924925 | standard |
| 7.3114 | 0.187855 | standard |
| 7.31171 | 0.187782 | standard |
| 7.31821 | 0.113927 | standard |
| 7.34066 | 0.12422 | standard |
| 7.34719 | 0.191906 | standard |
| 7.35376 | 0.0756474 | standard |
| 3.7175 | 1.0 | standard |
| 5.31866 | 0.789883 | standard |
| 7.22528 | 0.157556 | standard |
| 7.23097 | 0.178541 | standard |
| 7.2491 | 0.182129 | standard |
| 7.25462 | 0.203383 | standard |
| 7.30594 | 0.0944696 | standard |
| 7.31162 | 0.19486 | standard |
| 7.3173 | 0.11432 | standard |
| 7.34146 | 0.121518 | standard |
| 7.34699 | 0.187894 | standard |
| 7.35266 | 0.0797114 | standard |