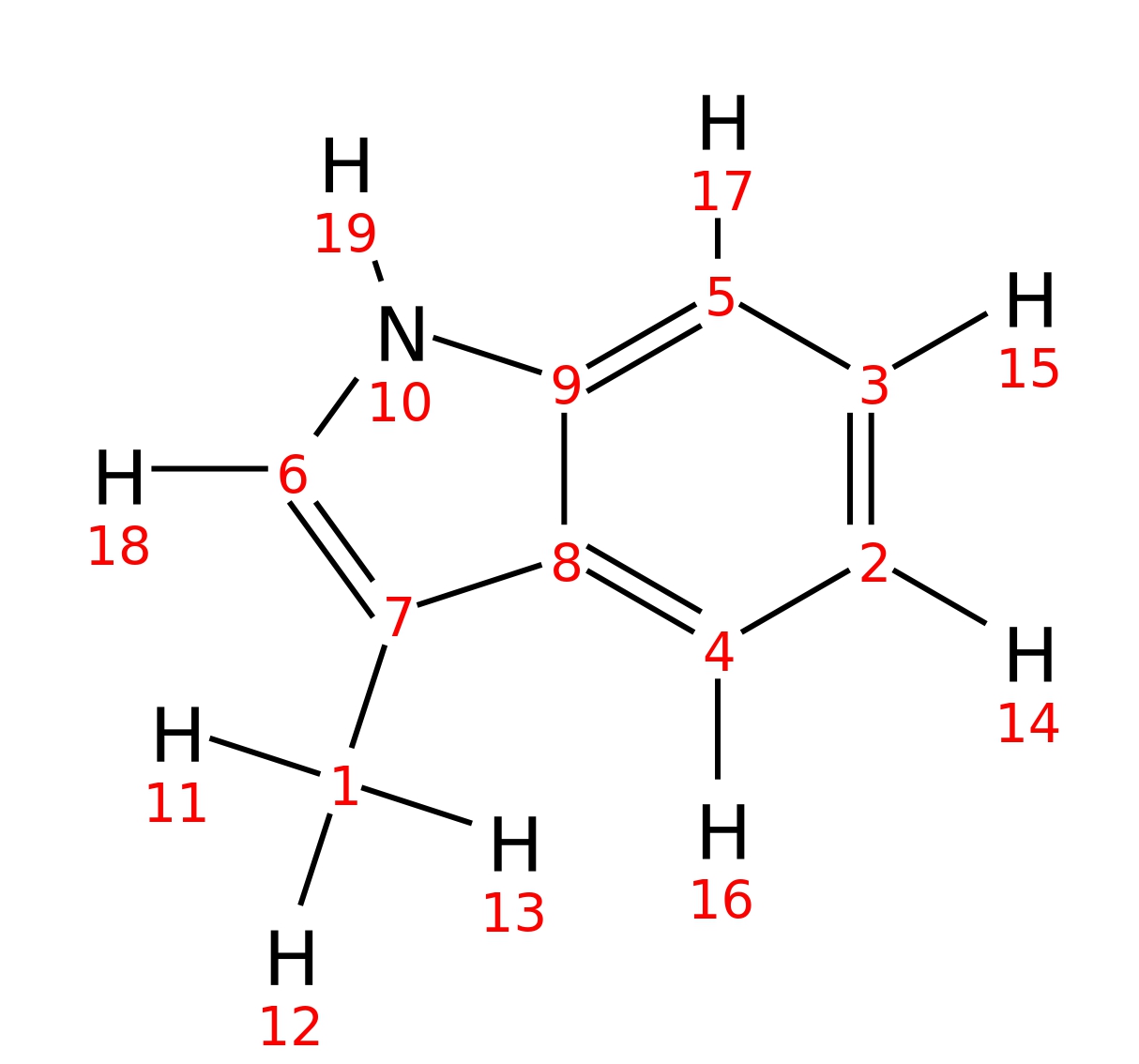
3-Methylindole
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01850 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H9N/c1-7-6-10-9-5-3-2-4-8(7)9/h2-6,10H,1H3 | |
| Note 1 | 15,17?14,16 | |
| Note 2 | RMSD .03374 - CDCl3 close in aromatic |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-methylindole | Solute | Saturated1 |
| CDCl3 | Solvent | 100% |
| TMS | Reference | 0.5% |
Spin System Matrix
| 11 | 12 | 13 | 18 | 16 | 14 | 15 | 17 | |
|---|---|---|---|---|---|---|---|---|
| 11 | 2.334 | -14.9 | -14.9 | 0.809 | 0 | 0 | 0 | 0 |
| 12 | 0 | 2.334 | -14.9 | 0.809 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 2.334 | 0.809 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 6.946 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 7.326 | 8.181 | 0 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 7.186 | 7.289 | 0 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 7.12 | 7.892 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.581 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.33405 | 1.0 | standard |
| 6.94622 | 0.304979 | standard |
| 7.10454 | 0.0777552 | standard |
| 7.1194 | 0.173957 | standard |
| 7.13458 | 0.121508 | standard |
| 7.17071 | 0.109617 | standard |
| 7.18638 | 0.172454 | standard |
| 7.20131 | 0.0900208 | standard |
| 7.31889 | 0.202037 | standard |
| 7.33502 | 0.162812 | standard |
| 7.57371 | 0.188066 | standard |
| 7.58929 | 0.179495 | standard |
| 2.33403 | 1.0 | standard |
| 6.94917 | 0.359995 | standard |
| 7.2246 | 0.557014 | standard |
| 7.50587 | 0.193417 | standard |
| 7.6854 | 0.0951461 | standard |
| 2.33405 | 1.0 | standard |
| 6.94685 | 0.327531 | standard |
| 7.08681 | 0.171951 | standard |
| 7.20156 | 0.41197 | standard |
| 7.28159 | 0.310962 | standard |
| 7.44368 | 0.0789191 | standard |
| 7.52822 | 0.168466 | standard |
| 7.58472 | 0.11206 | standard |
| 7.64949 | 0.108399 | standard |
| 2.33405 | 1.0 | standard |
| 6.94637 | 0.314436 | standard |
| 7.10042 | 0.181293 | standard |
| 7.14602 | 0.175428 | standard |
| 7.19233 | 0.330064 | standard |
| 7.28752 | 0.249942 | standard |
| 7.54068 | 0.163401 | standard |
| 7.58452 | 0.109374 | standard |
| 7.63144 | 0.116025 | standard |
| 2.33405 | 1.0 | standard |
| 6.94627 | 0.311602 | standard |
| 7.0266 | 0.0552901 | standard |
| 7.10573 | 0.179161 | standard |
| 7.14349 | 0.183639 | standard |
| 7.19048 | 0.292069 | standard |
| 7.29265 | 0.219333 | standard |
| 7.37385 | 0.0725611 | standard |
| 7.54477 | 0.162709 | standard |
| 7.57951 | 0.103215 | standard |
| 7.62553 | 0.119541 | standard |
| 2.33405 | 1.0 | standard |
| 6.94627 | 0.310735 | standard |
| 7.03647 | 0.051538 | standard |
| 7.10942 | 0.17205 | standard |
| 7.14432 | 0.194081 | standard |
| 7.1913 | 0.261941 | standard |
| 7.25936 | 0.148463 | standard |
| 7.29652 | 0.206166 | standard |
| 7.37345 | 0.0776051 | standard |
| 7.54807 | 0.163563 | standard |
| 7.62089 | 0.12315 | standard |
| 2.33405 | 1.0 | standard |
| 6.94616 | 0.30534 | standard |
| 7.07861 | 0.0578101 | standard |
| 7.11423 | 0.156471 | standard |
| 7.15109 | 0.254387 | standard |
| 7.18901 | 0.180286 | standard |
| 7.22457 | 0.0879 | standard |
| 7.27377 | 0.0213332 | standard |
| 7.31033 | 0.207471 | standard |
| 7.34827 | 0.126313 | standard |
| 7.56396 | 0.186068 | standard |
| 7.60044 | 0.160132 | standard |
| 2.33405 | 1.0 | standard |
| 6.94615 | 0.306635 | standard |
| 7.09323 | 0.0673188 | standard |
| 7.11751 | 0.168385 | standard |
| 7.1424 | 0.147004 | standard |
| 7.16154 | 0.128759 | standard |
| 7.18743 | 0.176003 | standard |
| 7.21175 | 0.0885287 | standard |
| 7.3147 | 0.211605 | standard |
| 7.34089 | 0.148636 | standard |
| 7.56913 | 0.193426 | standard |
| 7.59435 | 0.174985 | standard |
| 2.33405 | 1.0 | standard |
| 6.94618 | 0.304229 | standard |
| 7.10035 | 0.0728841 | standard |
| 7.11884 | 0.172273 | standard |
| 7.13764 | 0.129794 | standard |
| 7.1672 | 0.114811 | standard |
| 7.18673 | 0.173671 | standard |
| 7.20525 | 0.0890536 | standard |
| 7.31723 | 0.205569 | standard |
| 7.33722 | 0.157616 | standard |
| 7.57193 | 0.190801 | standard |
| 7.59119 | 0.177697 | standard |
| 2.33405 | 1.0 | standard |
| 6.94622 | 0.304928 | standard |
| 7.10458 | 0.0778611 | standard |
| 7.1194 | 0.173707 | standard |
| 7.13458 | 0.121327 | standard |
| 7.17071 | 0.109589 | standard |
| 7.18638 | 0.172437 | standard |
| 7.20131 | 0.0899249 | standard |
| 7.31889 | 0.201957 | standard |
| 7.33502 | 0.162671 | standard |
| 7.57376 | 0.188316 | standard |
| 7.58928 | 0.179507 | standard |
| 2.33405 | 1.0 | standard |
| 6.94618 | 0.306501 | standard |
| 7.10729 | 0.0815954 | standard |
| 7.11972 | 0.176345 | standard |
| 7.13235 | 0.117518 | standard |
| 7.17313 | 0.107481 | standard |
| 7.18615 | 0.172965 | standard |
| 7.19864 | 0.091282 | standard |
| 7.32006 | 0.200434 | standard |
| 7.33354 | 0.167726 | standard |
| 7.57495 | 0.187954 | standard |
| 7.58798 | 0.182666 | standard |
| 2.33405 | 1.0 | standard |
| 6.94622 | 0.30528 | standard |
| 7.10927 | 0.0842252 | standard |
| 7.11994 | 0.177004 | standard |
| 7.13076 | 0.113752 | standard |
| 7.17492 | 0.105619 | standard |
| 7.18606 | 0.171503 | standard |
| 7.19681 | 0.0920049 | standard |
| 7.32092 | 0.198508 | standard |
| 7.3325 | 0.17038 | standard |
| 7.57586 | 0.186947 | standard |
| 7.58701 | 0.183976 | standard |
| 2.33405 | 1.0 | standard |
| 6.94616 | 0.300751 | standard |
| 7.11001 | 0.0838561 | standard |
| 7.12 | 0.17578 | standard |
| 7.13012 | 0.110742 | standard |
| 7.17561 | 0.103475 | standard |
| 7.18599 | 0.170363 | standard |
| 7.19607 | 0.0908208 | standard |
| 7.32129 | 0.19504 | standard |
| 7.3321 | 0.16888 | standard |
| 7.57621 | 0.183667 | standard |
| 7.58664 | 0.181952 | standard |
| 2.33405 | 1.0 | standard |
| 6.94622 | 0.304381 | standard |
| 7.1107 | 0.0852353 | standard |
| 7.12005 | 0.176451 | standard |
| 7.12953 | 0.110462 | standard |
| 7.1762 | 0.10354 | standard |
| 7.18595 | 0.171051 | standard |
| 7.19543 | 0.0918545 | standard |
| 7.32159 | 0.195631 | standard |
| 7.33173 | 0.170888 | standard |
| 7.57655 | 0.18513 | standard |
| 7.58632 | 0.183213 | standard |
| 2.33409 | 1.00058 | standard |
| 6.94616 | 0.30511 | standard |
| 7.11184 | 0.0867513 | standard |
| 7.12016 | 0.177516 | standard |
| 7.12859 | 0.1088 | standard |
| 7.17724 | 0.102978 | standard |
| 7.18589 | 0.171985 | standard |
| 7.19434 | 0.0922472 | standard |
| 7.32214 | 0.194699 | standard |
| 7.33114 | 0.172841 | standard |
| 7.57705 | 0.184632 | standard |
| 7.58575 | 0.184505 | standard |
| 2.33405 | 1.0 | standard |
| 6.94616 | 0.300092 | standard |
| 7.11228 | 0.0867575 | standard |
| 7.12019 | 0.1763 | standard |
| 7.12819 | 0.107123 | standard |
| 7.17763 | 0.101583 | standard |
| 7.18589 | 0.16929 | standard |
| 7.19385 | 0.0919936 | standard |
| 7.32233 | 0.1926 | standard |
| 7.33089 | 0.171701 | standard |
| 7.57727 | 0.182755 | standard |
| 7.58555 | 0.183218 | standard |
| 2.33405 | 1.0 | standard |
| 6.94622 | 0.304034 | standard |
| 7.11267 | 0.0875639 | standard |
| 7.12022 | 0.177918 | standard |
| 7.1278 | 0.107355 | standard |
| 7.17803 | 0.101904 | standard |
| 7.18587 | 0.171773 | standard |
| 7.19345 | 0.0926503 | standard |
| 7.32253 | 0.193405 | standard |
| 7.33064 | 0.173564 | standard |
| 7.57749 | 0.18396 | standard |
| 7.58536 | 0.183988 | standard |
| 2.33405 | 1.0 | standard |
| 6.94616 | 0.310317 | standard |
| 7.11346 | 0.0895984 | standard |
| 7.12028 | 0.179562 | standard |
| 7.12715 | 0.107666 | standard |
| 7.17872 | 0.102682 | standard |
| 7.18583 | 0.173266 | standard |
| 7.19276 | 0.0941159 | standard |
| 7.32288 | 0.194663 | standard |
| 7.3303 | 0.176673 | standard |
| 7.57784 | 0.186317 | standard |
| 7.58496 | 0.186583 | standard |
| 2.33405 | 1.0 | standard |
| 6.94622 | 0.304333 | standard |
| 7.11455 | 0.0895338 | standard |
| 7.12031 | 0.176166 | standard |
| 7.12616 | 0.104262 | standard |
| 7.17981 | 0.100704 | standard |
| 7.18576 | 0.169937 | standard |
| 7.19168 | 0.0927226 | standard |
| 7.32342 | 0.190238 | standard |
| 7.32966 | 0.17622 | standard |
| 7.57833 | 0.183624 | standard |
| 7.58442 | 0.183308 | standard |