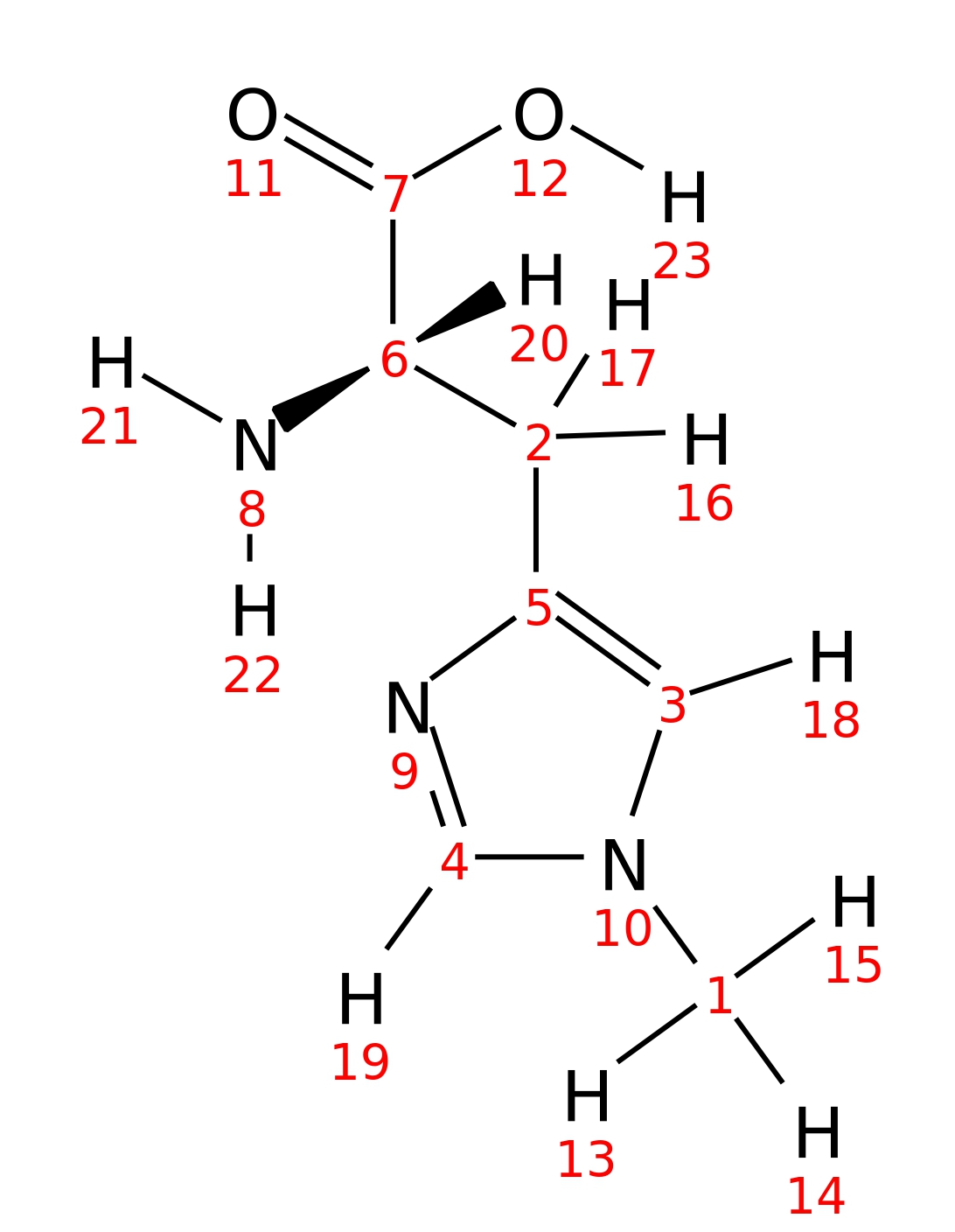
Ntau-Methyl-L-histidine
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.00931 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H11N3O2/c1-10-3-5(9-4-10)2-6(8)7(11)12/h3-4,6H,2,8H2,1H3,(H,11,12)/t6-/m0/s1 | |
| pKa | 1.69 | |
| pKa | 6.48 | |
| pKa | 8.85 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N(pai)-Methyl-L-histidine | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 13 | 14 | 15 | 19 | 18 | 16 | 17 | 20 | |
|---|---|---|---|---|---|---|---|---|
| 13 | 3.676 | -12.5 | -12.5 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 3.676 | -12.5 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 3.676 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 6.988 | 0.42 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 7.636 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 3.062 | -15.385 | 8.047 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 3.154 | 4.845 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.955 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.0358 | 0.0604018 | standard |
| 3.05178 | 0.0619348 | standard |
| 3.06664 | 0.114422 | standard |
| 3.08259 | 0.114525 | standard |
| 3.13591 | 0.117611 | standard |
| 3.14565 | 0.119551 | standard |
| 3.16664 | 0.0640912 | standard |
| 3.17638 | 0.0613703 | standard |
| 3.67604 | 1.0 | standard |
| 3.94271 | 0.0919932 | standard |
| 3.95259 | 0.10408 | standard |
| 3.9585 | 0.104073 | standard |
| 3.96838 | 0.0920234 | standard |
| 6.98846 | 0.325842 | standard |
| 7.63605 | 0.325842 | standard |
| 3.03495 | 0.208803 | standard |
| 3.17949 | 0.401146 | standard |
| 3.43377 | 0.0322041 | standard |
| 3.67609 | 1.0 | standard |
| 3.80827 | 0.180244 | standard |
| 3.95138 | 0.130303 | standard |
| 3.98426 | 0.120307 | standard |
| 4.12976 | 0.0694243 | standard |
| 6.98841 | 0.322363 | standard |
| 7.63604 | 0.322162 | standard |
| 2.77895 | 0.00995213 | standard |
| 3.03772 | 0.191989 | standard |
| 3.06557 | 0.202958 | standard |
| 3.15751 | 0.378101 | standard |
| 3.3214 | 0.0198951 | standard |
| 3.67604 | 1.0 | standard |
| 3.85541 | 0.127819 | standard |
| 3.95093 | 0.125823 | standard |
| 3.97156 | 0.120961 | standard |
| 4.06883 | 0.075389 | standard |
| 6.98841 | 0.323906 | standard |
| 7.63605 | 0.324112 | standard |
| 2.85671 | 0.0131351 | standard |
| 2.95153 | 0.0157311 | standard |
| 3.0497 | 0.176338 | standard |
| 3.08238 | 0.191283 | standard |
| 3.14555 | 0.331813 | standard |
| 3.27379 | 0.0207761 | standard |
| 3.34242 | 0.0121631 | standard |
| 3.67604 | 1.0 | standard |
| 3.87913 | 0.114097 | standard |
| 3.95014 | 0.122065 | standard |
| 3.96776 | 0.118456 | standard |
| 4.03937 | 0.078751 | standard |
| 6.98841 | 0.324596 | standard |
| 7.63605 | 0.324885 | standard |
| 2.88215 | 0.014731 | standard |
| 2.96631 | 0.0171032 | standard |
| 3.0535 | 0.169698 | standard |
| 3.08821 | 0.187271 | standard |
| 3.14101 | 0.297811 | standard |
| 3.25821 | 0.0217171 | standard |
| 3.31867 | 0.013057 | standard |
| 3.67604 | 1.0 | standard |
| 3.88728 | 0.110504 | standard |
| 3.94974 | 0.120046 | standard |
| 3.96675 | 0.116545 | standard |
| 4.0297 | 0.0799134 | standard |
| 6.98841 | 0.324485 | standard |
| 7.63605 | 0.324126 | standard |
| 2.90243 | 0.0165943 | standard |
| 2.97811 | 0.0186302 | standard |
| 3.05644 | 0.164022 | standard |
| 3.09302 | 0.185384 | standard |
| 3.13366 | 0.262402 | standard |
| 3.24582 | 0.0231713 | standard |
| 3.29987 | 0.0144021 | standard |
| 3.67604 | 1.0 | standard |
| 3.89383 | 0.107916 | standard |
| 3.94959 | 0.118518 | standard |
| 3.96591 | 0.115235 | standard |
| 4.02201 | 0.080925 | standard |
| 6.98846 | 0.324734 | standard |
| 7.63605 | 0.324725 | standard |
| 2.98962 | 0.0336263 | standard |
| 3.0284 | 0.0352265 | standard |
| 3.06648 | 0.137192 | standard |
| 3.10594 | 0.189072 | standard |
| 3.1152 | 0.182223 | standard |
| 3.1418 | 0.157973 | standard |
| 3.19309 | 0.03864 | standard |
| 3.21864 | 0.0320236 | standard |
| 3.67604 | 1.0 | standard |
| 3.92393 | 0.0974753 | standard |
| 3.94998 | 0.109083 | standard |
| 3.96204 | 0.106674 | standard |
| 3.98813 | 0.0870361 | standard |
| 6.98841 | 0.325032 | standard |
| 7.63605 | 0.324248 | standard |
| 3.01618 | 0.046063 | standard |
| 3.04249 | 0.0476982 | standard |
| 3.06744 | 0.126061 | standard |
| 3.09371 | 0.134696 | standard |
| 3.12631 | 0.132999 | standard |
| 3.14294 | 0.137063 | standard |
| 3.17755 | 0.0503228 | standard |
| 3.1942 | 0.0457334 | standard |
| 3.67604 | 1.0 | standard |
| 3.93431 | 0.0940533 | standard |
| 3.95112 | 0.105772 | standard |
| 3.96024 | 0.104297 | standard |
| 3.97707 | 0.0899151 | standard |
| 6.98846 | 0.324958 | standard |
| 7.63605 | 0.324958 | standard |
| 3.02867 | 0.0546598 | standard |
| 3.04851 | 0.0560559 | standard |
| 3.06714 | 0.119012 | standard |
| 3.08696 | 0.121839 | standard |
| 3.13215 | 0.123714 | standard |
| 3.14435 | 0.126102 | standard |
| 3.17052 | 0.0585199 | standard |
| 3.18282 | 0.055016 | standard |
| 3.67604 | 1.0 | standard |
| 3.93955 | 0.0924626 | standard |
| 3.95204 | 0.10465 | standard |
| 3.95914 | 0.103742 | standard |
| 3.97164 | 0.0914611 | standard |
| 6.98846 | 0.325891 | standard |
| 7.63605 | 0.32589 | standard |
| 3.0359 | 0.0604159 | standard |
| 3.05178 | 0.0619417 | standard |
| 3.06664 | 0.114193 | standard |
| 3.08259 | 0.114552 | standard |
| 3.13591 | 0.117578 | standard |
| 3.14565 | 0.119516 | standard |
| 3.16665 | 0.0639734 | standard |
| 3.17639 | 0.0613889 | standard |
| 3.67604 | 1.0 | standard |
| 3.94271 | 0.0918624 | standard |
| 3.95259 | 0.104037 | standard |
| 3.9585 | 0.10404 | standard |
| 3.96838 | 0.0920014 | standard |
| 6.98846 | 0.32567 | standard |
| 7.63605 | 0.32567 | standard |
| 3.04046 | 0.0642618 | standard |
| 3.05385 | 0.0661163 | standard |
| 3.06614 | 0.110493 | standard |
| 3.07942 | 0.110013 | standard |
| 3.13858 | 0.113358 | standard |
| 3.14665 | 0.115014 | standard |
| 3.16416 | 0.0681081 | standard |
| 3.17223 | 0.0658724 | standard |
| 3.67604 | 1.0 | standard |
| 3.94478 | 0.0920255 | standard |
| 3.95296 | 0.103498 | standard |
| 3.95803 | 0.103493 | standard |
| 3.96621 | 0.0920158 | standard |
| 6.98841 | 0.325086 | standard |
| 7.636 | 0.325086 | standard |
| 3.04373 | 0.0671518 | standard |
| 3.05524 | 0.0691919 | standard |
| 3.06574 | 0.107579 | standard |
| 3.07714 | 0.1067 | standard |
| 3.14046 | 0.110059 | standard |
| 3.14744 | 0.111525 | standard |
| 3.16247 | 0.0711075 | standard |
| 3.16936 | 0.0691763 | standard |
| 3.67604 | 1.0 | standard |
| 3.94626 | 0.0918897 | standard |
| 3.95335 | 0.103615 | standard |
| 3.95764 | 0.103716 | standard |
| 3.96463 | 0.0920698 | standard |
| 6.98846 | 0.325084 | standard |
| 7.63605 | 0.325084 | standard |
| 3.04502 | 0.0682588 | standard |
| 3.05573 | 0.0704336 | standard |
| 3.06555 | 0.106315 | standard |
| 3.07625 | 0.105165 | standard |
| 3.14135 | 0.108989 | standard |
| 3.14774 | 0.110316 | standard |
| 3.16178 | 0.0721593 | standard |
| 3.16827 | 0.0703665 | standard |
| 3.67604 | 1.0 | standard |
| 3.94695 | 0.0920706 | standard |
| 3.95344 | 0.103645 | standard |
| 3.95745 | 0.103539 | standard |
| 3.96404 | 0.0919266 | standard |
| 6.98846 | 0.323631 | standard |
| 7.63605 | 0.323739 | standard |
| 3.04611 | 0.0694099 | standard |
| 3.05613 | 0.0715385 | standard |
| 3.06535 | 0.105257 | standard |
| 3.07536 | 0.104179 | standard |
| 3.14205 | 0.107823 | standard |
| 3.14804 | 0.109192 | standard |
| 3.16128 | 0.0734279 | standard |
| 3.16728 | 0.0717202 | standard |
| 3.67604 | 1.0 | standard |
| 3.94745 | 0.0921053 | standard |
| 3.95353 | 0.103422 | standard |
| 3.95736 | 0.103382 | standard |
| 3.96344 | 0.0920993 | standard |
| 6.98846 | 0.322613 | standard |
| 7.63605 | 0.322613 | standard |
| 3.048 | 0.0709949 | standard |
| 3.05692 | 0.0734334 | standard |
| 3.06505 | 0.103374 | standard |
| 3.07397 | 0.102215 | standard |
| 3.14323 | 0.106037 | standard |
| 3.14854 | 0.107249 | standard |
| 3.16029 | 0.0748175 | standard |
| 3.1657 | 0.073554 | standard |
| 3.67604 | 1.0 | standard |
| 3.94834 | 0.09197 | standard |
| 3.95372 | 0.103195 | standard |
| 3.95717 | 0.1032 | standard |
| 3.96255 | 0.0921389 | standard |
| 6.98846 | 0.327128 | standard |
| 7.63605 | 0.327126 | standard |
| 3.04879 | 0.0719599 | standard |
| 3.05722 | 0.0742012 | standard |
| 3.06495 | 0.102808 | standard |
| 3.07337 | 0.101386 | standard |
| 3.14373 | 0.105005 | standard |
| 3.14884 | 0.106207 | standard |
| 3.15989 | 0.0759687 | standard |
| 3.165 | 0.0744205 | standard |
| 3.67604 | 1.0 | standard |
| 3.94873 | 0.0921728 | standard |
| 3.95381 | 0.103293 | standard |
| 3.95708 | 0.103296 | standard |
| 3.96216 | 0.0921776 | standard |
| 6.98846 | 0.326557 | standard |
| 7.63605 | 0.326557 | standard |
| 3.04938 | 0.0726209 | standard |
| 3.05742 | 0.0748301 | standard |
| 3.06485 | 0.102107 | standard |
| 3.07278 | 0.100624 | standard |
| 3.14422 | 0.104368 | standard |
| 3.14904 | 0.105524 | standard |
| 3.15959 | 0.0767217 | standard |
| 3.16441 | 0.0752117 | standard |
| 3.67604 | 1.0 | standard |
| 3.94903 | 0.0919495 | standard |
| 3.95391 | 0.103356 | standard |
| 3.95698 | 0.103357 | standard |
| 3.96186 | 0.0920244 | standard |
| 6.98846 | 0.325783 | standard |
| 7.63605 | 0.325786 | standard |
| 3.05057 | 0.0738078 | standard |
| 3.05791 | 0.0761432 | standard |
| 3.06455 | 0.100905 | standard |
| 3.07189 | 0.099283 | standard |
| 3.14501 | 0.103247 | standard |
| 3.14934 | 0.104334 | standard |
| 3.159 | 0.0780405 | standard |
| 3.16332 | 0.0766666 | standard |
| 3.67604 | 1.0 | standard |
| 3.94962 | 0.0921652 | standard |
| 3.95401 | 0.103533 | standard |
| 3.95678 | 0.103357 | standard |
| 3.96127 | 0.0918955 | standard |
| 6.98846 | 0.324276 | standard |
| 7.63605 | 0.324276 | standard |
| 3.05236 | 0.0756518 | standard |
| 3.05861 | 0.078009 | standard |
| 3.06425 | 0.099107 | standard |
| 3.0704 | 0.0973691 | standard |
| 3.1463 | 0.101353 | standard |
| 3.14994 | 0.102367 | standard |
| 3.1581 | 0.0798013 | standard |
| 3.16184 | 0.0785295 | standard |
| 3.67604 | 1.0 | standard |
| 3.95051 | 0.0919634 | standard |
| 3.9543 | 0.103866 | standard |
| 3.95659 | 0.103866 | standard |
| 3.96038 | 0.0919624 | standard |
| 6.98841 | 0.326227 | standard |
| 7.63605 | 0.320898 | standard |