D-Panthenol
Simulation outputs:
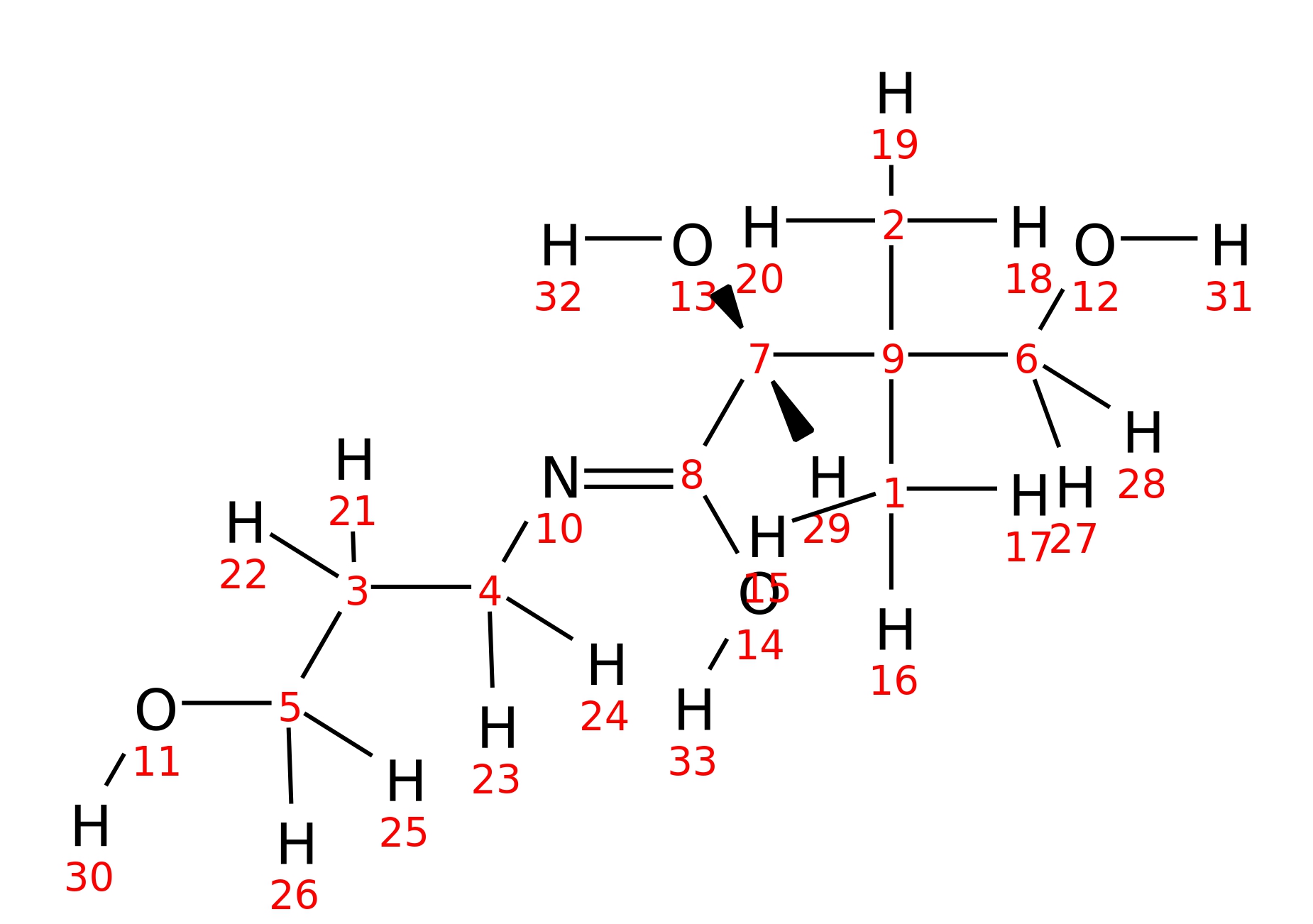
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01736 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H19NO4/c1-9(2,6-12)7(13)8(14)10-4-3-5-11/h7,11-13H,3-6H2,1-2H3,(H,10,14)/t7-/m0/s1 | |
| Note 1 | 15,16,17?18,19,20 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| D-Panthenol | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 27 | 28 | 29 | 23 | 24 | 21 | 22 | 25 | 26 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 0.895 | -12.5 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0.895 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0.895 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0.93 | -12.5 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0.93 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0.93 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 3.393 | -11.162 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.511 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.982 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.295 | -14.042 | 6.767 | 5.833 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.316 | 7.869 | 6.919 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.764 | -12.4 | 5.937 | 6.298 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.764 | 7.231 | 6.217 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.632 | -12.4 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.633 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 0.894852 | 1.0 | standard |
| 0.930161 | 0.999599 | standard |
| 1.73747 | 0.0470794 | standard |
| 1.75065 | 0.155237 | standard |
| 1.764 | 0.220623 | standard |
| 1.77734 | 0.15607 | standard |
| 1.79047 | 0.0471108 | standard |
| 3.26068 | 0.00906672 | standard |
| 3.27372 | 0.0205834 | standard |
| 3.28874 | 0.0952523 | standard |
| 3.29437 | 0.0996858 | standard |
| 3.30179 | 0.170476 | standard |
| 3.30871 | 0.172571 | standard |
| 3.31458 | 0.0986224 | standard |
| 3.32304 | 0.0896347 | standard |
| 3.33675 | 0.0209849 | standard |
| 3.35119 | 0.0121194 | standard |
| 3.38106 | 0.137266 | standard |
| 3.40347 | 0.198703 | standard |
| 3.50058 | 0.198591 | standard |
| 3.52289 | 0.136987 | standard |
| 3.61976 | 0.175215 | standard |
| 3.6326 | 0.322878 | standard |
| 3.64542 | 0.175207 | standard |
| 3.98163 | 0.332241 | standard |
| 0.900501 | 1.0 | standard |
| 0.924498 | 0.999717 | standard |
| 1.42942 | 0.0228501 | standard |
| 1.58663 | 0.0853983 | standard |
| 1.74979 | 0.152754 | standard |
| 1.91102 | 0.126855 | standard |
| 2.06828 | 0.0391861 | standard |
| 3.1462 | 0.154322 | standard |
| 3.31406 | 0.236725 | standard |
| 3.46399 | 0.533275 | standard |
| 3.64022 | 0.23733 | standard |
| 3.79823 | 0.111057 | standard |
| 3.98158 | 0.24921 | standard |
| 0.89614 | 1.0 | standard |
| 0.928871 | 0.99909 | standard |
| 1.53803 | 0.0286671 | standard |
| 1.64739 | 0.111143 | standard |
| 1.757 | 0.185615 | standard |
| 1.86637 | 0.146486 | standard |
| 1.97618 | 0.044565 | standard |
| 3.19596 | 0.165477 | standard |
| 3.25087 | 0.0625491 | standard |
| 3.30932 | 0.278969 | standard |
| 3.43433 | 0.403449 | standard |
| 3.46731 | 0.344536 | standard |
| 3.52844 | 0.192052 | standard |
| 3.63677 | 0.283288 | standard |
| 3.74208 | 0.132954 | standard |
| 3.98162 | 0.282916 | standard |
| 0.895304 | 1.00001 | standard |
| 0.929707 | 0.998418 | standard |
| 1.59501 | 0.033425 | standard |
| 1.6776 | 0.126089 | standard |
| 1.75993 | 0.20271 | standard |
| 1.84236 | 0.15539 | standard |
| 1.92503 | 0.0472141 | standard |
| 3.22226 | 0.172919 | standard |
| 3.30723 | 0.311424 | standard |
| 3.3946 | 0.162279 | standard |
| 3.43072 | 0.305802 | standard |
| 3.47314 | 0.297017 | standard |
| 3.55429 | 0.181112 | standard |
| 3.63455 | 0.300691 | standard |
| 3.71425 | 0.145273 | standard |
| 3.98163 | 0.300714 | standard |
| 0.895145 | 0.998986 | standard |
| 0.929869 | 1.0 | standard |
| 1.61407 | 0.035153 | standard |
| 1.68753 | 0.131157 | standard |
| 1.76075 | 0.207973 | standard |
| 1.83413 | 0.157877 | standard |
| 1.90769 | 0.0479615 | standard |
| 3.23122 | 0.174978 | standard |
| 3.30673 | 0.34475 | standard |
| 3.38346 | 0.1517 | standard |
| 3.42872 | 0.29151 | standard |
| 3.47525 | 0.286562 | standard |
| 3.56285 | 0.182157 | standard |
| 3.6012 | 0.0917575 | standard |
| 3.63428 | 0.301675 | standard |
| 3.70505 | 0.15008 | standard |
| 3.98163 | 0.306523 | standard |
| 0.895049 | 0.999762 | standard |
| 0.929964 | 1.0 | standard |
| 1.62928 | 0.0365125 | standard |
| 1.69545 | 0.135204 | standard |
| 1.76139 | 0.211781 | standard |
| 1.82747 | 0.159562 | standard |
| 1.89369 | 0.0485094 | standard |
| 3.23839 | 0.177248 | standard |
| 3.30726 | 0.337696 | standard |
| 3.37516 | 0.148003 | standard |
| 3.42681 | 0.282023 | standard |
| 3.4771 | 0.278781 | standard |
| 3.56989 | 0.188975 | standard |
| 3.63399 | 0.304159 | standard |
| 3.69766 | 0.153963 | standard |
| 3.98163 | 0.311033 | standard |
| 0.894865 | 1.00001 | standard |
| 0.930148 | 0.998334 | standard |
| 1.69723 | 0.0436209 | standard |
| 1.73033 | 0.154788 | standard |
| 1.76344 | 0.224312 | standard |
| 1.79654 | 0.158403 | standard |
| 1.82964 | 0.0484355 | standard |
| 3.20138 | 0.00323712 | standard |
| 3.23471 | 0.00899337 | standard |
| 3.27136 | 0.174788 | standard |
| 3.30558 | 0.299128 | standard |
| 3.33984 | 0.133327 | standard |
| 3.35899 | 0.109519 | standard |
| 3.41496 | 0.239472 | standard |
| 3.48907 | 0.236826 | standard |
| 3.54481 | 0.098423 | standard |
| 3.60075 | 0.175142 | standard |
| 3.63293 | 0.318167 | standard |
| 3.66488 | 0.170588 | standard |
| 3.98163 | 0.326863 | standard |
| 0.894854 | 0.998524 | standard |
| 0.930159 | 1.0 | standard |
| 1.71965 | 0.0463152 | standard |
| 1.74167 | 0.156923 | standard |
| 1.76381 | 0.225489 | standard |
| 1.78593 | 0.157714 | standard |
| 1.808 | 0.0469536 | standard |
| 3.23451 | 0.0047255 | standard |
| 3.2565 | 0.0119983 | standard |
| 3.28257 | 0.147008 | standard |
| 3.30397 | 0.22739 | standard |
| 3.30671 | 0.226963 | standard |
| 3.32529 | 0.10721 | standard |
| 3.33098 | 0.105212 | standard |
| 3.3546 | 0.0167084 | standard |
| 3.37184 | 0.12045 | standard |
| 3.40903 | 0.217273 | standard |
| 3.49492 | 0.216946 | standard |
| 3.53211 | 0.118045 | standard |
| 3.61129 | 0.174647 | standard |
| 3.63272 | 0.320619 | standard |
| 3.65404 | 0.174265 | standard |
| 3.98163 | 0.330273 | standard |
| 0.894852 | 1.0 | standard |
| 0.930161 | 0.999996 | standard |
| 1.73084 | 0.0468818 | standard |
| 1.7473 | 0.156641 | standard |
| 1.76394 | 0.223984 | standard |
| 1.7806 | 0.156915 | standard |
| 1.79706 | 0.0469943 | standard |
| 3.25099 | 0.00676157 | standard |
| 3.26736 | 0.0161713 | standard |
| 3.28634 | 0.109555 | standard |
| 3.29002 | 0.110724 | standard |
| 3.30254 | 0.187093 | standard |
| 3.30802 | 0.186653 | standard |
| 3.31867 | 0.0977991 | standard |
| 3.32598 | 0.0945099 | standard |
| 3.3432 | 0.0170813 | standard |
| 3.36135 | 0.012418 | standard |
| 3.37778 | 0.130333 | standard |
| 3.40565 | 0.205977 | standard |
| 3.4983 | 0.205786 | standard |
| 3.52626 | 0.129684 | standard |
| 3.61659 | 0.175107 | standard |
| 3.63265 | 0.322232 | standard |
| 3.64864 | 0.174866 | standard |
| 3.98163 | 0.331736 | standard |
| 0.894852 | 1.0 | standard |
| 0.930161 | 0.997263 | standard |
| 1.73747 | 0.0470214 | standard |
| 1.75065 | 0.155043 | standard |
| 1.764 | 0.220349 | standard |
| 1.77732 | 0.156105 | standard |
| 1.79047 | 0.0470447 | standard |
| 3.26068 | 0.00905872 | standard |
| 3.27372 | 0.0205644 | standard |
| 3.28874 | 0.0951283 | standard |
| 3.29437 | 0.0995508 | standard |
| 3.30179 | 0.170255 | standard |
| 3.30871 | 0.172347 | standard |
| 3.31458 | 0.0984873 | standard |
| 3.32304 | 0.0895226 | standard |
| 3.33675 | 0.0209639 | standard |
| 3.35119 | 0.0121035 | standard |
| 3.38116 | 0.137126 | standard |
| 3.40347 | 0.19847 | standard |
| 3.50058 | 0.198349 | standard |
| 3.52289 | 0.136835 | standard |
| 3.61976 | 0.175005 | standard |
| 3.6326 | 0.322486 | standard |
| 3.64542 | 0.174997 | standard |
| 3.98163 | 0.331853 | standard |
| 0.894852 | 1.0 | standard |
| 0.930162 | 0.998874 | standard |
| 1.74193 | 0.0470595 | standard |
| 1.75288 | 0.155213 | standard |
| 1.76403 | 0.218635 | standard |
| 1.77519 | 0.154976 | standard |
| 1.78611 | 0.0470777 | standard |
| 3.26702 | 0.0114395 | standard |
| 3.27784 | 0.0250252 | standard |
| 3.29037 | 0.0892084 | standard |
| 3.29732 | 0.104332 | standard |
| 3.30118 | 0.165707 | standard |
| 3.30932 | 0.17401 | standard |
| 3.32116 | 0.087035 | standard |
| 3.3326 | 0.0252511 | standard |
| 3.34464 | 0.0139209 | standard |
| 3.38334 | 0.142142 | standard |
| 3.40188 | 0.193686 | standard |
| 3.50206 | 0.193587 | standard |
| 3.52071 | 0.141951 | standard |
| 3.62189 | 0.17534 | standard |
| 3.6326 | 0.322572 | standard |
| 3.64324 | 0.175267 | standard |
| 3.98163 | 0.332433 | standard |
| 0.894852 | 1.0 | standard |
| 0.930162 | 0.999848 | standard |
| 1.74515 | 0.0472398 | standard |
| 1.75449 | 0.154143 | standard |
| 1.76406 | 0.216735 | standard |
| 1.77364 | 0.153853 | standard |
| 1.78299 | 0.0472208 | standard |
| 3.27148 | 0.0137563 | standard |
| 3.2807 | 0.029309 | standard |
| 3.29145 | 0.0855257 | standard |
| 3.30051 | 0.187268 | standard |
| 3.30975 | 0.21544 | standard |
| 3.31997 | 0.0852377 | standard |
| 3.32973 | 0.0294309 | standard |
| 3.34008 | 0.0158931 | standard |
| 3.38483 | 0.145775 | standard |
| 3.40079 | 0.190177 | standard |
| 3.50325 | 0.190126 | standard |
| 3.51912 | 0.145676 | standard |
| 3.62338 | 0.175431 | standard |
| 3.63258 | 0.323386 | standard |
| 3.64176 | 0.175288 | standard |
| 3.98163 | 0.332783 | standard |
| 0.894852 | 0.999201 | standard |
| 0.930162 | 1.0 | standard |
| 1.74644 | 0.0472433 | standard |
| 1.75513 | 0.154264 | standard |
| 1.76407 | 0.215529 | standard |
| 1.77297 | 0.154332 | standard |
| 1.7817 | 0.0472145 | standard |
| 3.27326 | 0.0148613 | standard |
| 3.28181 | 0.0312754 | standard |
| 3.29194 | 0.0837939 | standard |
| 3.30043 | 0.213155 | standard |
| 3.30966 | 0.1974 | standard |
| 3.31948 | 0.0850708 | standard |
| 3.32858 | 0.0314241 | standard |
| 3.3383 | 0.0168371 | standard |
| 3.38542 | 0.147179 | standard |
| 3.4003 | 0.18867 | standard |
| 3.50365 | 0.188613 | standard |
| 3.51853 | 0.147079 | standard |
| 3.62402 | 0.175216 | standard |
| 3.63258 | 0.324125 | standard |
| 3.64111 | 0.175528 | standard |
| 3.98163 | 0.332743 | standard |
| 0.894852 | 1.0 | standard |
| 0.930162 | 0.99973 | standard |
| 1.74753 | 0.0472286 | standard |
| 1.7557 | 0.153095 | standard |
| 1.76408 | 0.216196 | standard |
| 1.77248 | 0.153465 | standard |
| 1.78061 | 0.0472534 | standard |
| 3.27474 | 0.0159212 | standard |
| 3.28277 | 0.0332692 | standard |
| 3.29224 | 0.0826413 | standard |
| 3.30052 | 0.199451 | standard |
| 3.30997 | 0.166259 | standard |
| 3.31908 | 0.0845152 | standard |
| 3.32762 | 0.0333661 | standard |
| 3.33671 | 0.0177711 | standard |
| 3.38592 | 0.148481 | standard |
| 3.3999 | 0.187441 | standard |
| 3.50405 | 0.18739 | standard |
| 3.51803 | 0.148395 | standard |
| 3.62457 | 0.175536 | standard |
| 3.63258 | 0.32218 | standard |
| 3.64057 | 0.175359 | standard |
| 3.98163 | 0.332886 | standard |
| 0.894852 | 1.0 | standard |
| 0.930162 | 0.999066 | standard |
| 1.74936 | 0.0473047 | standard |
| 1.7566 | 0.152172 | standard |
| 1.76406 | 0.212077 | standard |
| 1.77153 | 0.153073 | standard |
| 1.77878 | 0.0472956 | standard |
| 3.27722 | 0.0178982 | standard |
| 3.28436 | 0.0368897 | standard |
| 3.2928 | 0.080435 | standard |
| 3.29999 | 0.150634 | standard |
| 3.30212 | 0.100537 | standard |
| 3.30711 | 0.0870208 | standard |
| 3.31042 | 0.144261 | standard |
| 3.31848 | 0.0833542 | standard |
| 3.32604 | 0.0369496 | standard |
| 3.33413 | 0.0195031 | standard |
| 3.38681 | 0.150568 | standard |
| 3.39921 | 0.185255 | standard |
| 3.50474 | 0.185214 | standard |
| 3.51714 | 0.150504 | standard |
| 3.62546 | 0.175501 | standard |
| 3.63258 | 0.324453 | standard |
| 3.6397 | 0.175286 | standard |
| 3.98163 | 0.33286 | standard |
| 0.894852 | 0.999013 | standard |
| 0.930162 | 1.0 | standard |
| 1.7501 | 0.0471769 | standard |
| 1.75699 | 0.151653 | standard |
| 1.76408 | 0.21389 | standard |
| 1.77117 | 0.152481 | standard |
| 1.77804 | 0.0471928 | standard |
| 3.27831 | 0.0188493 | standard |
| 3.28499 | 0.0385153 | standard |
| 3.29303 | 0.0791001 | standard |
| 3.29978 | 0.142874 | standard |
| 3.30286 | 0.0890866 | standard |
| 3.30647 | 0.082658 | standard |
| 3.31063 | 0.139699 | standard |
| 3.31818 | 0.0828607 | standard |
| 3.32539 | 0.0385451 | standard |
| 3.33304 | 0.0203161 | standard |
| 3.38721 | 0.151475 | standard |
| 3.39891 | 0.184358 | standard |
| 3.50504 | 0.184301 | standard |
| 3.51684 | 0.151394 | standard |
| 3.62586 | 0.175541 | standard |
| 3.63258 | 0.324216 | standard |
| 3.63933 | 0.175539 | standard |
| 3.98163 | 0.332884 | standard |
| 0.894851 | 1.0 | standard |
| 0.930162 | 0.999653 | standard |
| 1.75085 | 0.0473397 | standard |
| 1.75734 | 0.151811 | standard |
| 1.76407 | 0.215367 | standard |
| 1.77082 | 0.152611 | standard |
| 1.77734 | 0.0471957 | standard |
| 3.2792 | 0.0197293 | standard |
| 3.28556 | 0.0401742 | standard |
| 3.29312 | 0.079257 | standard |
| 3.29962 | 0.1385 | standard |
| 3.30352 | 0.0866205 | standard |
| 3.30593 | 0.0833056 | standard |
| 3.31075 | 0.136438 | standard |
| 3.31798 | 0.0823209 | standard |
| 3.32481 | 0.040175 | standard |
| 3.33215 | 0.021075 | standard |
| 3.3875 | 0.152338 | standard |
| 3.39861 | 0.183607 | standard |
| 3.50534 | 0.183573 | standard |
| 3.51644 | 0.152288 | standard |
| 3.62615 | 0.175513 | standard |
| 3.63258 | 0.322948 | standard |
| 3.63898 | 0.175215 | standard |
| 3.98163 | 0.333018 | standard |
| 0.894851 | 1.0 | standard |
| 0.930162 | 0.999763 | standard |
| 1.75204 | 0.0473095 | standard |
| 1.75798 | 0.152807 | standard |
| 1.76409 | 0.21057 | standard |
| 1.77019 | 0.150594 | standard |
| 1.77611 | 0.0473458 | standard |
| 3.28079 | 0.0213693 | standard |
| 3.28654 | 0.0431961 | standard |
| 3.29342 | 0.0779344 | standard |
| 3.29932 | 0.133024 | standard |
| 3.30472 | 0.117901 | standard |
| 3.31105 | 0.131729 | standard |
| 3.31768 | 0.0812072 | standard |
| 3.32383 | 0.0432016 | standard |
| 3.33046 | 0.0225241 | standard |
| 3.388 | 0.153721 | standard |
| 3.39821 | 0.182183 | standard |
| 3.50583 | 0.182178 | standard |
| 3.51595 | 0.153708 | standard |
| 3.62675 | 0.175572 | standard |
| 3.63258 | 0.321499 | standard |
| 3.63839 | 0.175183 | standard |
| 3.98163 | 0.333081 | standard |
| 0.894851 | 1.0 | standard |
| 0.930162 | 0.998579 | standard |
| 1.75392 | 0.047342 | standard |
| 1.75889 | 0.150435 | standard |
| 1.76411 | 0.211401 | standard |
| 1.76926 | 0.152119 | standard |
| 1.77427 | 0.0471423 | standard |
| 3.28317 | 0.0241434 | standard |
| 3.28802 | 0.0483009 | standard |
| 3.29382 | 0.0750294 | standard |
| 3.29882 | 0.126027 | standard |
| 3.30371 | 0.0721616 | standard |
| 3.30585 | 0.0726722 | standard |
| 3.31155 | 0.125183 | standard |
| 3.31708 | 0.0807812 | standard |
| 3.32236 | 0.0481048 | standard |
| 3.32799 | 0.0249571 | standard |
| 3.38889 | 0.15584 | standard |
| 3.39742 | 0.179937 | standard |
| 3.50653 | 0.179916 | standard |
| 3.51506 | 0.155812 | standard |
| 3.62764 | 0.175451 | standard |
| 3.63258 | 0.324076 | standard |
| 3.6375 | 0.174877 | standard |
| 3.98163 | 0.332944 | standard |