4-Hydroxyphenylacetonitrile
Simulation outputs:
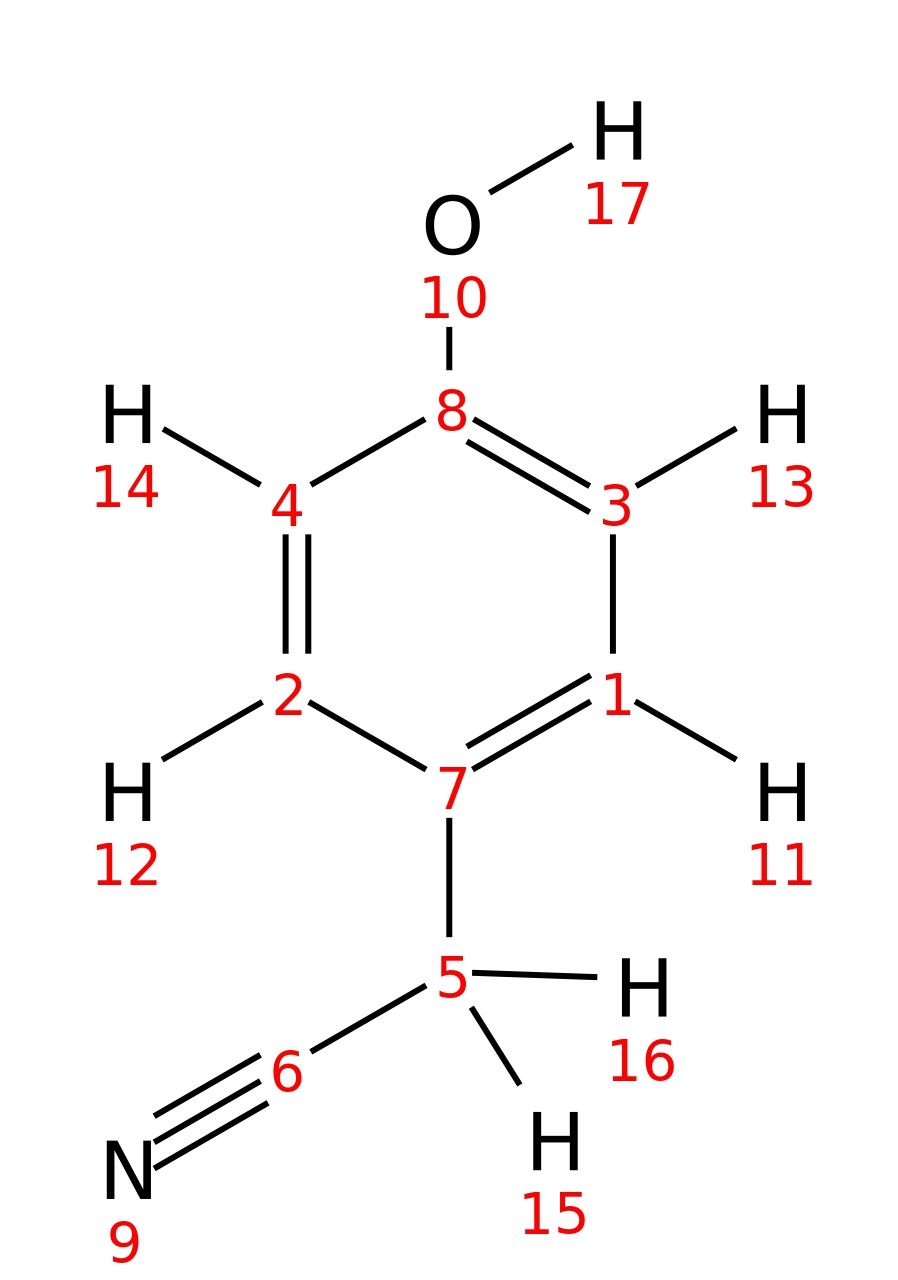
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01443 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H7NO/c9-6-5-7-1-3-8(10)4-2-7/h1-4,10H,5H2 | |
| Note 1 | 11,12?13,14 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4-Hydroxyphenylacetonitrile | Solute | SaturatedmM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 13 | 11 | 12 | 14 | 15 | 16 | |
|---|---|---|---|---|---|---|
| 13 | 7.264 | 8.529 | 0 | 2.416 | 1.0 | 1.0 |
| 11 | 0 | 6.906 | 3.255 | 0 | 0 | 0 |
| 12 | 0 | 0 | 6.906 | 8.529 | 0 | 0 |
| 14 | 0 | 0 | 0 | 7.264 | 1.0 | 1.0 |
| 15 | 0 | 0 | 0 | 0 | 3.818 | -12.4 |
| 16 | 0 | 0 | 0 | 0 | 0 | 3.818 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.81767 | 1.0 | standard |
| 6.89739 | 0.48947 | standard |
| 6.91429 | 0.536157 | standard |
| 7.25613 | 0.430785 | standard |
| 7.27264 | 0.394057 | standard |
| -0.978332 | 2.125e-06 | standard |
| 3.81725 | 1.0 | standard |
| 6.76731 | 0.242387 | standard |
| 6.8198 | 0.149394 | standard |
| 6.98575 | 0.775795 | standard |
| 7.1846 | 0.627295 | standard |
| 7.40118 | 0.206028 | standard |
| -0.933897 | 1.125e-06 | standard |
| 3.81751 | 1.0 | standard |
| 6.82054 | 0.324074 | standard |
| 6.96419 | 0.693961 | standard |
| 7.20678 | 0.55597 | standard |
| 7.34816 | 0.26687 | standard |
| 3.81759 | 1.0 | standard |
| 6.84484 | 0.368899 | standard |
| 6.95175 | 0.652228 | standard |
| 7.21925 | 0.52166 | standard |
| 7.32413 | 0.300473 | standard |
| 3.81763 | 1.00001 | standard |
| 6.85249 | 0.384286 | standard |
| 6.94729 | 0.635277 | standard |
| 7.22369 | 0.507843 | standard |
| 7.3166 | 0.312455 | standard |
| 3.81763 | 1.00001 | standard |
| 6.85847 | 0.396497 | standard |
| 6.94364 | 0.622412 | standard |
| 7.2273 | 0.498889 | standard |
| 7.31083 | 0.322059 | standard |
| 3.81767 | 1.0 | standard |
| 6.88361 | 0.456632 | standard |
| 6.92594 | 0.571123 | standard |
| 7.24472 | 0.458786 | standard |
| 7.28614 | 0.369206 | standard |
| 3.81769 | 1.00001 | standard |
| 6.8914 | 0.475789 | standard |
| 6.91958 | 0.552104 | standard |
| 7.25098 | 0.443212 | standard |
| 7.27856 | 0.383802 | standard |
| 3.81768 | 1.0 | standard |
| 6.89514 | 0.484773 | standard |
| 6.91626 | 0.542867 | standard |
| 7.25421 | 0.436887 | standard |
| 7.27487 | 0.391196 | standard |
| 3.81767 | 1.0 | standard |
| 6.89739 | 0.489497 | standard |
| 6.91429 | 0.536171 | standard |
| 7.25612 | 0.430199 | standard |
| 7.27265 | 0.394909 | standard |
| 3.81768 | 1.0 | standard |
| 6.89887 | 0.50063 | standard |
| 6.91291 | 0.527011 | standard |
| 7.25749 | 0.423397 | standard |
| 7.27122 | 0.408834 | standard |
| 3.81769 | 1.00007 | standard |
| 6.89985 | 0.503833 | standard |
| 6.91194 | 0.528337 | standard |
| 7.25846 | 0.425819 | standard |
| 7.27027 | 0.406774 | standard |
| 3.81769 | 1.00008 | standard |
| 6.90034 | 0.506872 | standard |
| 6.91154 | 0.528081 | standard |
| 7.25884 | 0.423894 | standard |
| 7.26988 | 0.405148 | standard |
| 3.81769 | 1.00009 | standard |
| 6.90064 | 0.505299 | standard |
| 6.91124 | 0.525125 | standard |
| 7.25909 | 0.420636 | standard |
| 7.26951 | 0.406864 | standard |
| 3.81767 | 1.0 | standard |
| 6.90123 | 0.508938 | standard |
| 6.91066 | 0.516102 | standard |
| 7.2597 | 0.412917 | standard |
| 7.2689 | 0.410594 | standard |
| 3.81769 | 1.00012 | standard |
| 6.90152 | 0.510077 | standard |
| 6.91045 | 0.513883 | standard |
| 7.26 | 0.41527 | standard |
| 7.26861 | 0.412483 | standard |
| 3.81767 | 1.0 | standard |
| 6.90172 | 0.513285 | standard |
| 6.91017 | 0.512663 | standard |
| 7.26017 | 0.413919 | standard |
| 7.26843 | 0.4134 | standard |
| 3.81767 | 1.0 | standard |
| 6.90211 | 0.505942 | standard |
| 6.90987 | 0.509765 | standard |
| 7.26056 | 0.411854 | standard |
| 7.26804 | 0.412496 | standard |
| 3.81769 | 1.00022 | standard |
| 6.9028 | 0.506751 | standard |
| 6.90928 | 0.510207 | standard |
| 7.26116 | 0.41406 | standard |
| 7.26749 | 0.411349 | standard |