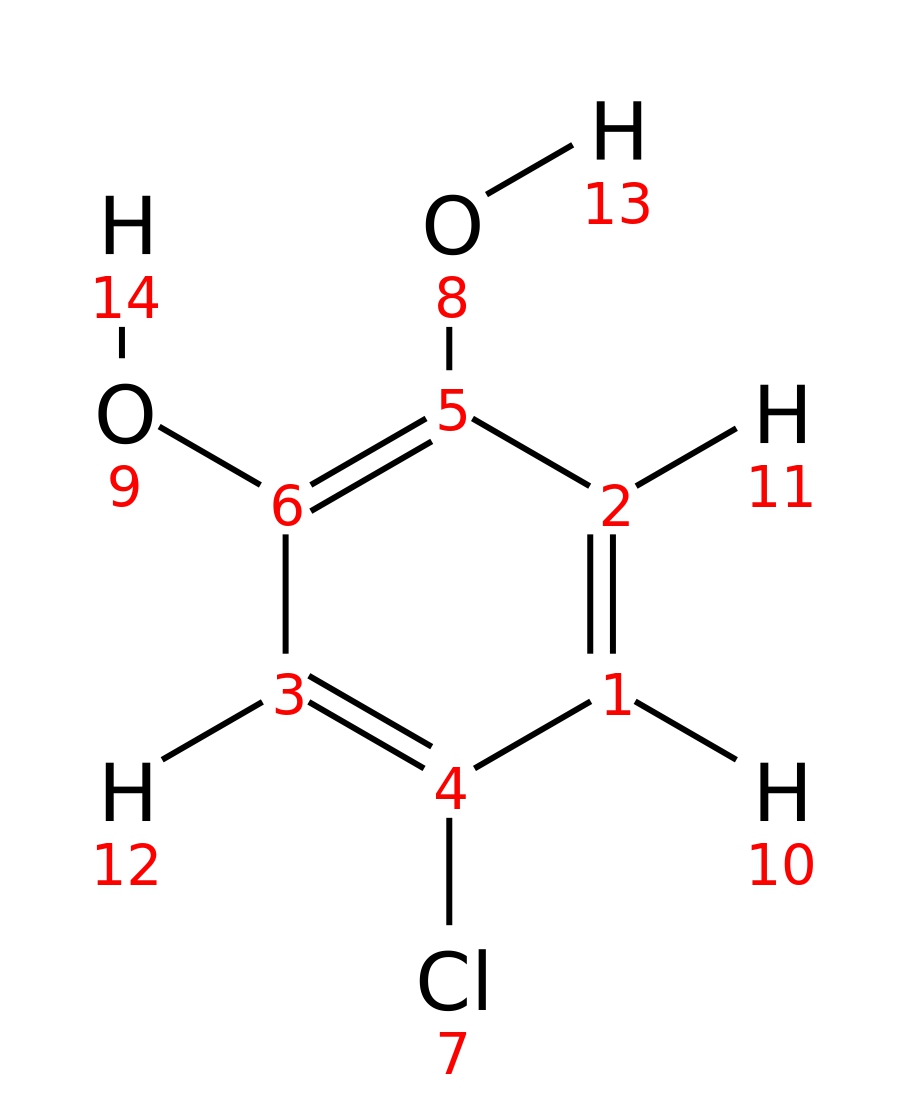
4-Chlorocatechol
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.03030 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H5ClO2/c7-4-1-2-5(8)6(9)3-4/h1-3,8-9H | |
| Note 1 | intensity of 12 is ~.5 expected intensity (relaxation?) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4-Chlorocatechol | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 10 | 11 | 12 | |
|---|---|---|---|
| 10 | 6.821 | 8.89 | 2.966 |
| 11 | 0 | 6.855 | 0 |
| 12 | 0 | 0 | 6.935 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 6.80692 | 0.212 | standard |
| 6.81237 | 0.222425 | standard |
| 6.82468 | 0.52915 | standard |
| 6.83011 | 0.589102 | standard |
| 6.84846 | 1.00058 | standard |
| 6.8662 | 0.376634 | standard |
| 6.93278 | 0.773204 | standard |
| 6.93825 | 0.711246 | standard |
| -0.965041 | 7.125e-06 | standard |
| 6.85075 | 1.0 | standard |
| 6.90629 | 0.522436 | standard |
| -0.833123 | 3.125e-06 | standard |
| 6.67379 | 0.0195751 | standard |
| 6.84649 | 1.0 | standard |
| 6.93319 | 0.470715 | standard |
| -0.860697 | 2.125e-06 | standard |
| 6.71178 | 0.0226952 | standard |
| 6.84405 | 1.0 | standard |
| 6.93006 | 0.468691 | standard |
| -0.445007 | 1.125e-06 | standard |
| 6.72493 | 0.02485 | standard |
| 6.84324 | 1.0 | standard |
| 6.92604 | 0.453055 | standard |
| -0.526737 | 1.125e-06 | standard |
| 6.73544 | 0.027458 | standard |
| 6.76167 | 0.0297641 | standard |
| 6.84328 | 1.0 | standard |
| 6.92369 | 0.429176 | standard |
| 6.78144 | 0.074868 | standard |
| 6.79326 | 0.074011 | standard |
| 6.82588 | 0.497287 | standard |
| 6.84301 | 1.00003 | standard |
| 6.88707 | 0.13119 | standard |
| 6.9302 | 0.548147 | standard |
| 6.94145 | 0.467426 | standard |
| 6.79599 | 0.131548 | standard |
| 6.80453 | 0.134477 | standard |
| 6.82557 | 0.533639 | standard |
| 6.83413 | 0.661861 | standard |
| 6.84593 | 1.0 | standard |
| 6.8753 | 0.22158 | standard |
| 6.93141 | 0.677801 | standard |
| 6.93982 | 0.603881 | standard |
| 6.80288 | 0.176743 | standard |
| 6.80959 | 0.183863 | standard |
| 6.8251 | 0.534328 | standard |
| 6.8318 | 0.616014 | standard |
| 6.84738 | 1.00024 | standard |
| 6.86949 | 0.306318 | standard |
| 6.9322 | 0.739711 | standard |
| 6.93887 | 0.670713 | standard |
| 6.80692 | 0.212043 | standard |
| 6.81237 | 0.222467 | standard |
| 6.82468 | 0.529129 | standard |
| 6.83011 | 0.589065 | standard |
| 6.84846 | 1.00058 | standard |
| 6.8662 | 0.376736 | standard |
| 6.93278 | 0.773221 | standard |
| 6.93825 | 0.711277 | standard |
| 6.80939 | 0.243536 | standard |
| 6.81407 | 0.256558 | standard |
| 6.82426 | 0.530865 | standard |
| 6.82893 | 0.578603 | standard |
| 6.84931 | 1.0 | standard |
| 6.86407 | 0.443028 | standard |
| 6.93316 | 0.80663 | standard |
| 6.93782 | 0.75286 | standard |
| 6.81116 | 0.270313 | standard |
| 6.81526 | 0.285256 | standard |
| 6.82385 | 0.532892 | standard |
| 6.82794 | 0.571944 | standard |
| 6.84995 | 1.00126 | standard |
| 6.86264 | 0.495292 | standard |
| 6.93345 | 0.829932 | standard |
| 6.93749 | 0.784853 | standard |
| 6.81196 | 0.284226 | standard |
| 6.81565 | 0.299335 | standard |
| 6.82375 | 0.535327 | standard |
| 6.82755 | 0.570985 | standard |
| 6.85025 | 1.00144 | standard |
| 6.86214 | 0.518068 | standard |
| 6.9336 | 0.845701 | standard |
| 6.93734 | 0.796492 | standard |
| 6.81255 | 0.295188 | standard |
| 6.81605 | 0.310821 | standard |
| 6.82365 | 0.538529 | standard |
| 6.82715 | 0.571141 | standard |
| 6.85055 | 1.00163 | standard |
| 6.86165 | 0.5392 | standard |
| 6.9337 | 0.856757 | standard |
| 6.93719 | 0.813172 | standard |
| 6.81353 | 0.315788 | standard |
| 6.81665 | 0.332121 | standard |
| 6.82335 | 0.539054 | standard |
| 6.82656 | 0.566788 | standard |
| 6.85094 | 1.00206 | standard |
| 6.86081 | 0.577933 | standard |
| 6.93389 | 0.873862 | standard |
| 6.93705 | 0.835674 | standard |
| 6.81393 | 0.323826 | standard |
| 6.81695 | 0.340686 | standard |
| 6.82325 | 0.541332 | standard |
| 6.82627 | 0.566901 | standard |
| 6.85114 | 1.0023 | standard |
| 6.86051 | 0.594783 | standard |
| 6.93394 | 0.883595 | standard |
| 6.93696 | 0.845123 | standard |
| 6.81432 | 0.331482 | standard |
| 6.81715 | 0.3482 | standard |
| 6.82314 | 0.538142 | standard |
| 6.82607 | 0.561789 | standard |
| 6.85129 | 1.0 | standard |
| 6.86021 | 0.607215 | standard |
| 6.93404 | 0.889165 | standard |
| 6.93686 | 0.852516 | standard |
| 6.81491 | 0.345282 | standard |
| 6.81755 | 0.362552 | standard |
| 6.82294 | 0.540971 | standard |
| 6.82558 | 0.56129 | standard |
| 6.85159 | 1.0 | standard |
| 6.85972 | 0.634765 | standard |
| 6.93413 | 0.899781 | standard |
| 6.93676 | 0.865665 | standard |
| 6.8159 | 0.379055 | standard |
| 6.81804 | 0.382729 | standard |
| 6.82274 | 0.547057 | standard |
| 6.82488 | 0.562867 | standard |
| 6.85208 | 1.0 | standard |
| 6.85892 | 0.680652 | standard |
| 6.93433 | 0.914888 | standard |
| 6.93657 | 0.898812 | standard |