R-Pantolactone
Simulation outputs:
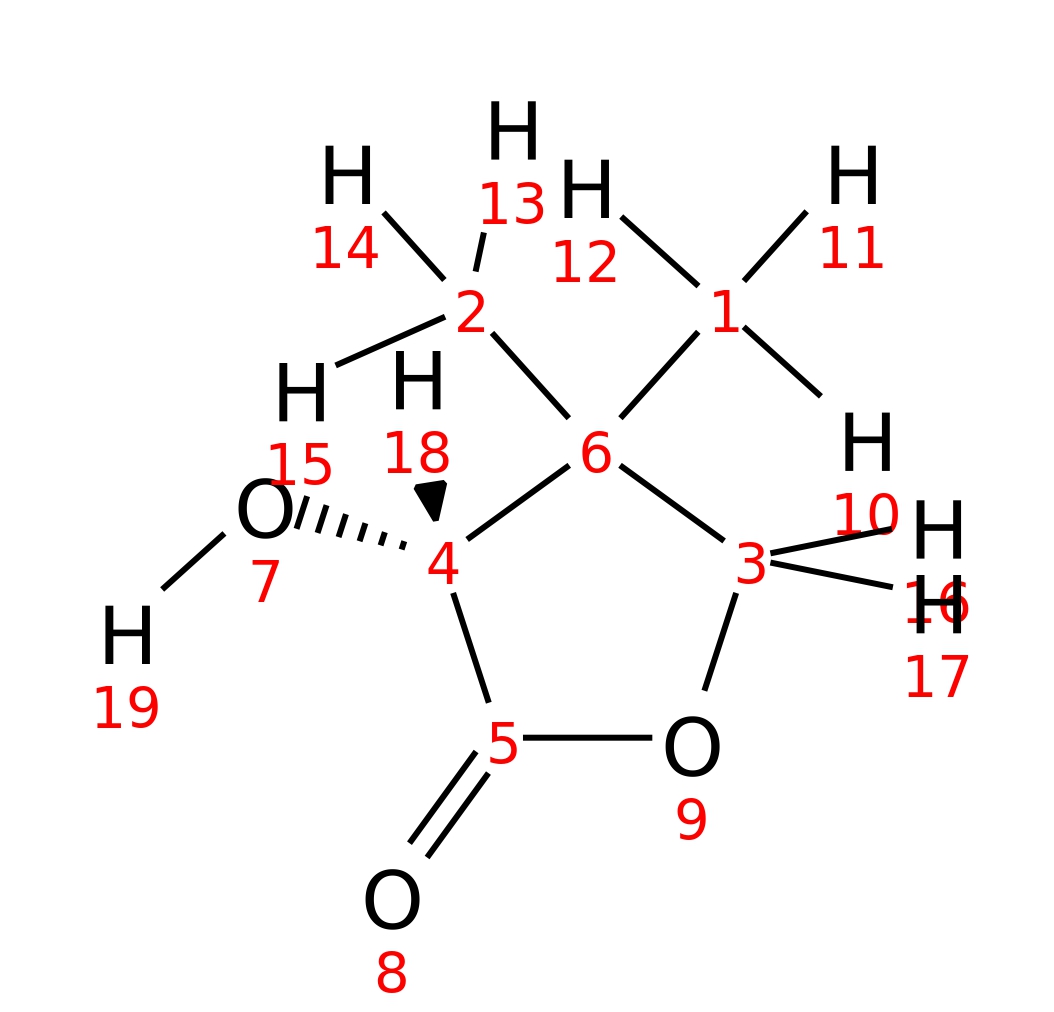
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01090 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H10O3/c1-6(2)3-9-5(8)4(6)7/h4,7H,3H2,1-2H3/t4-/m0/s1 | |
| Note 1 | 10,11,12?13,14,15 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| (R)-(-)-Pantolactone | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 10 | 11 | 12 | 18 | 16 | 17 | 13 | 14 | 15 | |
|---|---|---|---|---|---|---|---|---|---|
| 10 | 1.012 | -12.5 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 1.012 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 1.012 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 4.373 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 4.136 | -9.048 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 4.092 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 1.184 | -12.5 | -12.5 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.184 | -12.5 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.184 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.01179 | 1.0 | standard |
| 1.18428 | 0.999619 | standard |
| 4.08073 | 0.10645 | standard |
| 4.09887 | 0.232225 | standard |
| 4.12853 | 0.232584 | standard |
| 4.14667 | 0.106584 | standard |
| 4.37272 | 0.332003 | standard |
| 1.0119 | 1.0 | standard |
| 1.18417 | 0.999057 | standard |
| 4.11371 | 0.643188 | standard |
| 4.37266 | 0.331328 | standard |
| 1.01182 | 0.99988 | standard |
| 1.18425 | 1.0 | standard |
| 4.11365 | 0.62871 | standard |
| 4.37271 | 0.331468 | standard |
| 1.0118 | 0.99986 | standard |
| 1.18427 | 1.0 | standard |
| 3.99844 | 0.0179743 | standard |
| 4.11365 | 0.588071 | standard |
| 4.22969 | 0.020632 | standard |
| 4.37272 | 0.331178 | standard |
| 1.0118 | 1.0 | standard |
| 1.18427 | 0.999795 | standard |
| 4.00965 | 0.0205881 | standard |
| 4.11365 | 0.556803 | standard |
| 4.21794 | 0.0220574 | standard |
| 4.37272 | 0.331004 | standard |
| 1.0118 | 1.0 | standard |
| 1.18427 | 0.998867 | standard |
| 4.01897 | 0.0231854 | standard |
| 4.11371 | 0.521436 | standard |
| 4.20852 | 0.0241413 | standard |
| 4.37272 | 0.331541 | standard |
| 1.01179 | 1.0 | standard |
| 1.18428 | 0.999533 | standard |
| 4.05948 | 0.0523812 | standard |
| 4.10476 | 0.311356 | standard |
| 4.12253 | 0.311125 | standard |
| 4.16781 | 0.0522292 | standard |
| 4.37272 | 0.331604 | standard |
| 1.01179 | 1.0 | standard |
| 1.18428 | 0.999369 | standard |
| 4.07182 | 0.0770104 | standard |
| 4.10205 | 0.268321 | standard |
| 4.12534 | 0.267946 | standard |
| 4.15547 | 0.0769374 | standard |
| 4.37272 | 0.331424 | standard |
| 1.01179 | 1.0 | standard |
| 1.18428 | 0.999742 | standard |
| 4.07756 | 0.0939296 | standard |
| 4.10016 | 0.246171 | standard |
| 4.12723 | 0.24615 | standard |
| 4.14983 | 0.0941926 | standard |
| 4.37272 | 0.331922 | standard |
| 1.01179 | 1.0 | standard |
| 1.18428 | 0.999827 | standard |
| 4.08073 | 0.106741 | standard |
| 4.09887 | 0.232322 | standard |
| 4.12853 | 0.232465 | standard |
| 4.14657 | 0.106678 | standard |
| 4.37272 | 0.332179 | standard |
| 1.01179 | 0.999496 | standard |
| 1.18428 | 1.0 | standard |
| 4.08281 | 0.115963 | standard |
| 4.09787 | 0.222593 | standard |
| 4.12952 | 0.222468 | standard |
| 4.14459 | 0.115659 | standard |
| 4.37272 | 0.331568 | standard |
| 1.01179 | 0.999717 | standard |
| 1.18428 | 1.0 | standard |
| 4.08419 | 0.122817 | standard |
| 4.09708 | 0.215233 | standard |
| 4.13021 | 0.214953 | standard |
| 4.1432 | 0.122812 | standard |
| 4.37272 | 0.331949 | standard |
| 1.01179 | 1.0 | standard |
| 1.18428 | 0.999328 | standard |
| 4.08469 | 0.125498 | standard |
| 4.09678 | 0.212302 | standard |
| 4.13061 | 0.21264 | standard |
| 4.1426 | 0.125215 | standard |
| 4.37272 | 0.332184 | standard |
| 1.01179 | 0.999844 | standard |
| 1.18428 | 1.0 | standard |
| 4.08518 | 0.127738 | standard |
| 4.09649 | 0.209462 | standard |
| 4.13081 | 0.209431 | standard |
| 4.14221 | 0.127692 | standard |
| 4.37272 | 0.331357 | standard |
| 1.01179 | 1.0 | standard |
| 1.18428 | 0.999908 | standard |
| 4.08598 | 0.131878 | standard |
| 4.09599 | 0.205341 | standard |
| 4.1313 | 0.204867 | standard |
| 4.14142 | 0.131597 | standard |
| 4.37272 | 0.331905 | standard |
| 1.01179 | 1.0 | standard |
| 1.18428 | 0.999027 | standard |
| 4.08627 | 0.134069 | standard |
| 4.09579 | 0.203131 | standard |
| 4.1315 | 0.203373 | standard |
| 4.14102 | 0.13397 | standard |
| 4.37272 | 0.331764 | standard |
| 1.01179 | 1.0 | standard |
| 1.18428 | 0.999952 | standard |
| 4.08657 | 0.135667 | standard |
| 4.09559 | 0.201719 | standard |
| 4.1317 | 0.201856 | standard |
| 4.14082 | 0.135751 | standard |
| 4.37272 | 0.331976 | standard |
| 1.01179 | 1.0 | standard |
| 1.18428 | 0.999689 | standard |
| 4.08707 | 0.137989 | standard |
| 4.0953 | 0.198371 | standard |
| 4.1321 | 0.198278 | standard |
| 4.14032 | 0.138379 | standard |
| 4.37272 | 0.331483 | standard |
| 1.01179 | 1.0 | standard |
| 1.18428 | 0.999525 | standard |
| 4.08786 | 0.142966 | standard |
| 4.0948 | 0.193853 | standard |
| 4.13259 | 0.193718 | standard |
| 4.13953 | 0.142806 | standard |
| 4.37272 | 0.331316 | standard |