Cis-3-chloroacrylic-acid
Simulation outputs:
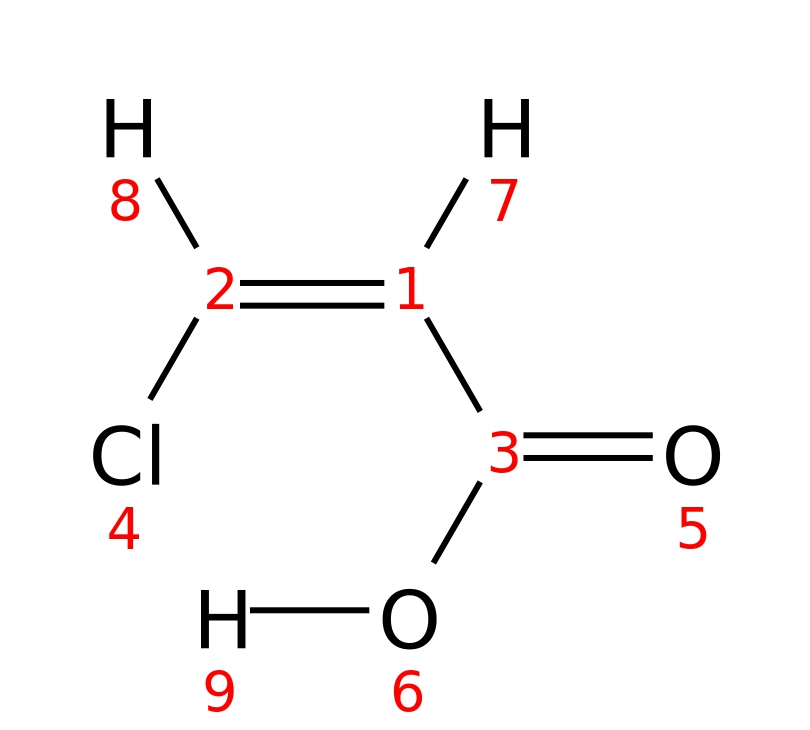
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.02024 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C3H3ClO2/c4-2-1-3(5)6/h1-2H,(H,5,6)/b2-1- | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| cis-3-chloroacrylic acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 7 | 8 | |
|---|---|---|
| 7 | 6.303 | 7.659 |
| 8 | 0 | 6.375 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 6.29189 | 0.605029 | standard |
| 6.31102 | 1.0 | standard |
| 6.36716 | 1.0 | standard |
| 6.38629 | 0.605029 | standard |
| -0.948477 | 2.125e-06 | standard |
| 6.14404 | 0.0351841 | standard |
| 6.33914 | 1.0 | standard |
| 6.53414 | 0.0352011 | standard |
| -0.76528 | 1.125e-06 | standard |
| 6.20269 | 0.066645 | standard |
| 6.33944 | 1.0 | standard |
| 6.47549 | 0.066645 | standard |
| -0.87012 | 1.125e-06 | standard |
| 6.23144 | 0.11611 | standard |
| 6.32807 | 0.999999 | standard |
| 6.35021 | 1.0 | standard |
| 6.44674 | 0.116153 | standard |
| -0.927945 | 1.125e-06 | standard |
| 6.24086 | 0.141735 | standard |
| 6.32631 | 1.0 | standard |
| 6.35197 | 1.0 | standard |
| 6.43742 | 0.141735 | standard |
| -0.987854 | 1.125e-06 | standard |
| 6.24823 | 0.167052 | standard |
| 6.32494 | 1.00001 | standard |
| 6.35334 | 1.00001 | standard |
| 6.43005 | 0.167052 | standard |
| 6.27892 | 0.377935 | standard |
| 6.31717 | 1.0 | standard |
| 6.36111 | 1.0 | standard |
| 6.39936 | 0.377935 | standard |
| 6.28783 | 0.514774 | standard |
| 6.3133 | 1.0 | standard |
| 6.36488 | 0.99994 | standard |
| 6.39045 | 0.514643 | standard |
| 6.29189 | 0.60494 | standard |
| 6.31102 | 1.0 | standard |
| 6.36716 | 1.0 | standard |
| 6.38629 | 0.60494 | standard |
| 6.29426 | 0.667566 | standard |
| 6.30963 | 0.999865 | standard |
| 6.36865 | 1.0 | standard |
| 6.38392 | 0.667778 | standard |
| 6.29575 | 0.713596 | standard |
| 6.30854 | 1.0 | standard |
| 6.36964 | 1.0 | standard |
| 6.38243 | 0.713596 | standard |
| 6.29684 | 0.748592 | standard |
| 6.30774 | 1.0 | standard |
| 6.37044 | 1.0 | standard |
| 6.38134 | 0.748592 | standard |
| 6.29724 | 0.763028 | standard |
| 6.30745 | 1.0 | standard |
| 6.37073 | 1.0 | standard |
| 6.38094 | 0.763028 | standard |
| 6.29763 | 0.776028 | standard |
| 6.30715 | 1.0 | standard |
| 6.37103 | 1.0 | standard |
| 6.38055 | 0.776028 | standard |
| 6.29823 | 0.797747 | standard |
| 6.30675 | 0.9997 | standard |
| 6.37153 | 1.0 | standard |
| 6.37995 | 0.79813 | standard |
| 6.29843 | 0.807148 | standard |
| 6.30656 | 0.999685 | standard |
| 6.37172 | 1.0 | standard |
| 6.37975 | 0.807545 | standard |
| 6.29872 | 0.816179 | standard |
| 6.30636 | 1.0 | standard |
| 6.37182 | 0.999666 | standard |
| 6.37956 | 0.815761 | standard |
| 6.29912 | 0.831336 | standard |
| 6.30606 | 1.0 | standard |
| 6.37222 | 1.0 | standard |
| 6.37916 | 0.831336 | standard |
| 6.29962 | 0.855121 | standard |
| 6.30556 | 1.0 | standard |
| 6.37262 | 1.0 | standard |
| 6.37856 | 0.855121 | standard |