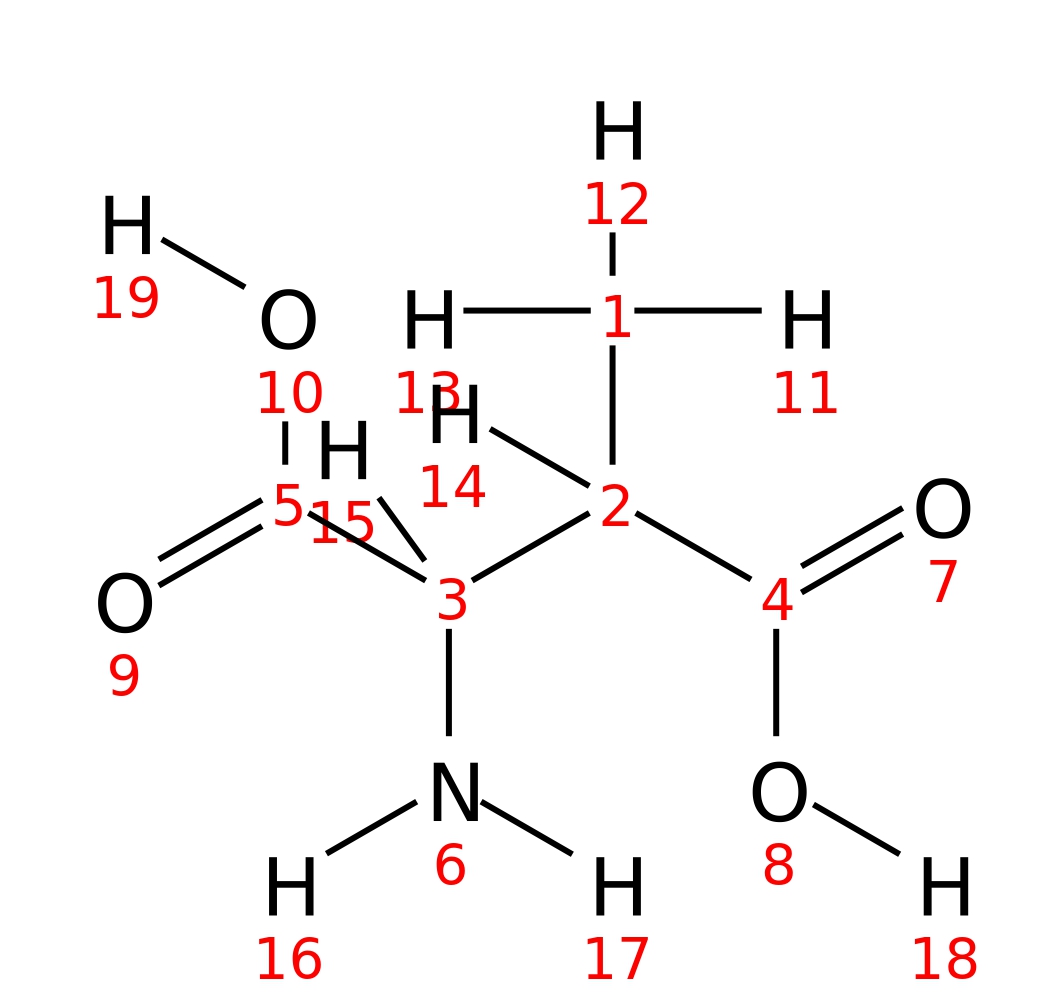
DL-threo-beta-Methylaspartate
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01030 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C5H9NO4/c1-2(4(7)8)3(6)5(9)10/h2-3H,6H2,1H3,(H,7,8)(H,9,10)/t2-,3+/m0/s1 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| DL-threo-beta-Methylaspartate | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 11 | 12 | 13 | 14 | 15 | |
|---|---|---|---|---|---|
| 11 | 1.127 | -12.5 | -12.5 | 7.693 | 0 |
| 12 | 0 | 1.127 | -12.5 | 7.693 | 0 |
| 13 | 0 | 0 | 1.127 | 7.693 | 0 |
| 14 | 0 | 0 | 0 | 2.933 | 3.141 |
| 15 | 0 | 0 | 0 | 0 | 3.992 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.11914 | 1.00021 | standard |
| 1.13456 | 1.00021 | standard |
| 2.90712 | 0.0487811 | standard |
| 2.91342 | 0.0538563 | standard |
| 2.92254 | 0.140105 | standard |
| 2.9287 | 0.14323 | standard |
| 2.93789 | 0.143226 | standard |
| 2.94405 | 0.140085 | standard |
| 2.95324 | 0.0538595 | standard |
| 2.95944 | 0.0489048 | standard |
| 3.98942 | 0.358619 | standard |
| 3.99556 | 0.358619 | standard |
| 1.02701 | 0.782398 | standard |
| 1.21714 | 1.0 | standard |
| 2.62273 | 0.0616321 | standard |
| 2.6996 | 0.0710812 | standard |
| 2.80503 | 0.114666 | standard |
| 2.88097 | 0.133814 | standard |
| 2.99696 | 0.0984634 | standard |
| 3.07333 | 0.111238 | standard |
| 3.19384 | 0.0375911 | standard |
| 3.2716 | 0.0394761 | standard |
| 3.95566 | 0.364069 | standard |
| 4.03215 | 0.320997 | standard |
| 1.06082 | 0.85976 | standard |
| 1.18848 | 1.00001 | standard |
| 2.72221 | 0.0560885 | standard |
| 2.77385 | 0.064238 | standard |
| 2.84816 | 0.123908 | standard |
| 2.89899 | 0.135875 | standard |
| 2.976 | 0.111948 | standard |
| 3.02701 | 0.119135 | standard |
| 3.10467 | 0.0418531 | standard |
| 3.1566 | 0.041164 | standard |
| 3.96748 | 0.358163 | standard |
| 4.01874 | 0.330528 | standard |
| 1.07767 | 0.896412 | standard |
| 1.1736 | 1.00001 | standard |
| 2.77361 | 0.0537842 | standard |
| 2.81242 | 0.061213 | standard |
| 2.86906 | 0.129507 | standard |
| 2.90734 | 0.138919 | standard |
| 2.96487 | 0.12062 | standard |
| 3.00326 | 0.124314 | standard |
| 3.06086 | 0.0443421 | standard |
| 3.09981 | 0.0425145 | standard |
| 3.9736 | 0.356105 | standard |
| 4.01211 | 0.336471 | standard |
| 1.08323 | 0.911077 | standard |
| 1.16853 | 1.0 | standard |
| 2.79092 | 0.0531542 | standard |
| 2.8254 | 0.0603231 | standard |
| 2.87604 | 0.131832 | standard |
| 2.91001 | 0.138692 | standard |
| 2.96117 | 0.125218 | standard |
| 2.99531 | 0.126196 | standard |
| 3.04641 | 0.045278 | standard |
| 3.08102 | 0.043099 | standard |
| 3.97568 | 0.354135 | standard |
| 4.00992 | 0.341384 | standard |
| 1.08767 | 0.919126 | standard |
| 1.16447 | 1.00001 | standard |
| 2.80487 | 0.0525851 | standard |
| 2.83591 | 0.0595761 | standard |
| 2.88152 | 0.133892 | standard |
| 2.91218 | 0.139562 | standard |
| 2.95819 | 0.127129 | standard |
| 2.98898 | 0.127678 | standard |
| 3.03485 | 0.0459969 | standard |
| 3.06598 | 0.0435144 | standard |
| 3.97731 | 0.353839 | standard |
| 4.00814 | 0.342811 | standard |
| 1.10748 | 0.965435 | standard |
| 1.14588 | 1.0 | standard |
| 2.86845 | 0.0511244 | standard |
| 2.88398 | 0.0560745 | standard |
| 2.90681 | 0.137375 | standard |
| 2.92233 | 0.140653 | standard |
| 2.9452 | 0.138878 | standard |
| 2.96067 | 0.135393 | standard |
| 2.98355 | 0.0503594 | standard |
| 2.9991 | 0.0454652 | standard |
| 3.98484 | 0.353201 | standard |
| 4.00024 | 0.353201 | standard |
| 1.11396 | 0.992784 | standard |
| 1.13964 | 1.00001 | standard |
| 2.88991 | 0.0487803 | standard |
| 2.90024 | 0.0537166 | standard |
| 2.91546 | 0.137142 | standard |
| 2.92583 | 0.14037 | standard |
| 2.94109 | 0.143806 | standard |
| 2.95146 | 0.141068 | standard |
| 2.96661 | 0.0538884 | standard |
| 2.97705 | 0.0487671 | standard |
| 3.98737 | 0.356911 | standard |
| 3.99771 | 0.357262 | standard |
| 1.11723 | 1.0 | standard |
| 1.13646 | 0.99904 | standard |
| 2.90069 | 0.0488852 | standard |
| 2.90846 | 0.0538779 | standard |
| 2.91991 | 0.139882 | standard |
| 2.92763 | 0.143033 | standard |
| 2.93912 | 0.142959 | standard |
| 2.94685 | 0.138944 | standard |
| 2.95819 | 0.0538776 | standard |
| 2.96607 | 0.0487845 | standard |
| 3.98864 | 0.358357 | standard |
| 3.99634 | 0.357468 | standard |
| 1.11914 | 1.00021 | standard |
| 1.13456 | 1.00021 | standard |
| 2.90712 | 0.0487769 | standard |
| 2.91342 | 0.0538491 | standard |
| 2.92254 | 0.140096 | standard |
| 2.9287 | 0.143219 | standard |
| 2.93789 | 0.143215 | standard |
| 2.94405 | 0.140076 | standard |
| 2.95324 | 0.0538526 | standard |
| 2.95944 | 0.0489008 | standard |
| 3.98942 | 0.358605 | standard |
| 3.99556 | 0.358605 | standard |
| 1.12048 | 1.00016 | standard |
| 1.13327 | 1.00016 | standard |
| 2.91147 | 0.0488303 | standard |
| 2.91669 | 0.0538798 | standard |
| 2.92428 | 0.140063 | standard |
| 2.92947 | 0.143237 | standard |
| 2.93709 | 0.143065 | standard |
| 2.94224 | 0.140195 | standard |
| 2.94987 | 0.0538324 | standard |
| 2.95509 | 0.0488193 | standard |
| 3.98991 | 0.35808 | standard |
| 3.99507 | 0.358082 | standard |
| 1.12137 | 1.00023 | standard |
| 1.13238 | 1.00022 | standard |
| 2.91453 | 0.0487412 | standard |
| 2.91906 | 0.0538548 | standard |
| 2.92556 | 0.140279 | standard |
| 2.92997 | 0.143357 | standard |
| 2.93656 | 0.143354 | standard |
| 2.94096 | 0.140272 | standard |
| 2.9475 | 0.0539289 | standard |
| 2.95192 | 0.0489325 | standard |
| 3.9903 | 0.35876 | standard |
| 3.99468 | 0.35876 | standard |
| 1.12177 | 0.998969 | standard |
| 1.132 | 1.0 | standard |
| 2.91582 | 0.048867 | standard |
| 2.91995 | 0.0538498 | standard |
| 2.92606 | 0.140065 | standard |
| 2.93017 | 0.143133 | standard |
| 2.93633 | 0.14314 | standard |
| 2.94047 | 0.139932 | standard |
| 2.9465 | 0.0539877 | standard |
| 2.95064 | 0.0489015 | standard |
| 3.99049 | 0.358318 | standard |
| 3.99459 | 0.358318 | standard |
| 1.12209 | 1.0 | standard |
| 1.13168 | 0.998823 | standard |
| 2.91691 | 0.0487532 | standard |
| 2.92085 | 0.0538727 | standard |
| 2.92652 | 0.140284 | standard |
| 2.93037 | 0.14318 | standard |
| 2.93613 | 0.143353 | standard |
| 2.93997 | 0.140073 | standard |
| 2.94571 | 0.05388 | standard |
| 2.94955 | 0.0489538 | standard |
| 3.99059 | 0.358852 | standard |
| 3.99439 | 0.358852 | standard |
| 1.12259 | 1.0 | standard |
| 1.13114 | 0.999608 | standard |
| 2.91869 | 0.0489015 | standard |
| 2.92213 | 0.0538364 | standard |
| 2.92724 | 0.140047 | standard |
| 2.93067 | 0.142179 | standard |
| 2.93576 | 0.142531 | standard |
| 2.93925 | 0.139943 | standard |
| 2.94433 | 0.0538958 | standard |
| 2.94777 | 0.0489138 | standard |
| 3.99078 | 0.358385 | standard |
| 3.9942 | 0.358709 | standard |
| 1.12281 | 0.998334 | standard |
| 1.13091 | 1.0 | standard |
| 2.91948 | 0.0489168 | standard |
| 2.92273 | 0.0538866 | standard |
| 2.92754 | 0.139666 | standard |
| 2.93083 | 0.14309 | standard |
| 2.93566 | 0.143224 | standard |
| 2.93889 | 0.140101 | standard |
| 2.94373 | 0.0538311 | standard |
| 2.94698 | 0.0489067 | standard |
| 3.99088 | 0.358614 | standard |
| 3.9941 | 0.358617 | standard |
| 1.12306 | 0.997653 | standard |
| 1.13071 | 1.0 | standard |
| 2.92017 | 0.0487635 | standard |
| 2.92332 | 0.0539343 | standard |
| 2.92783 | 0.139833 | standard |
| 2.93093 | 0.142755 | standard |
| 2.93553 | 0.143426 | standard |
| 2.93859 | 0.140018 | standard |
| 2.94324 | 0.0537699 | standard |
| 2.94628 | 0.0489725 | standard |
| 3.99098 | 0.356906 | standard |
| 3.9941 | 0.357375 | standard |
| 1.12338 | 1.0 | standard |
| 1.13039 | 0.99727 | standard |
| 2.92136 | 0.0487923 | standard |
| 2.92422 | 0.0539126 | standard |
| 2.92833 | 0.139306 | standard |
| 2.93113 | 0.143154 | standard |
| 2.93533 | 0.143081 | standard |
| 2.93817 | 0.140043 | standard |
| 2.94224 | 0.0540295 | standard |
| 2.9451 | 0.0488226 | standard |
| 3.99108 | 0.357548 | standard |
| 3.9939 | 0.357089 | standard |
| 1.1239 | 0.997511 | standard |
| 1.12982 | 1.0 | standard |
| 2.92314 | 0.0486452 | standard |
| 2.9256 | 0.0538281 | standard |
| 2.92908 | 0.140264 | standard |
| 2.93146 | 0.142834 | standard |
| 2.93503 | 0.143575 | standard |
| 2.93737 | 0.140568 | standard |
| 2.94086 | 0.0538562 | standard |
| 2.94332 | 0.0486562 | standard |
| 3.99137 | 0.359124 | standard |
| 3.99371 | 0.359125 | standard |