6-Hydroxynicotinic-acid
Simulation outputs:
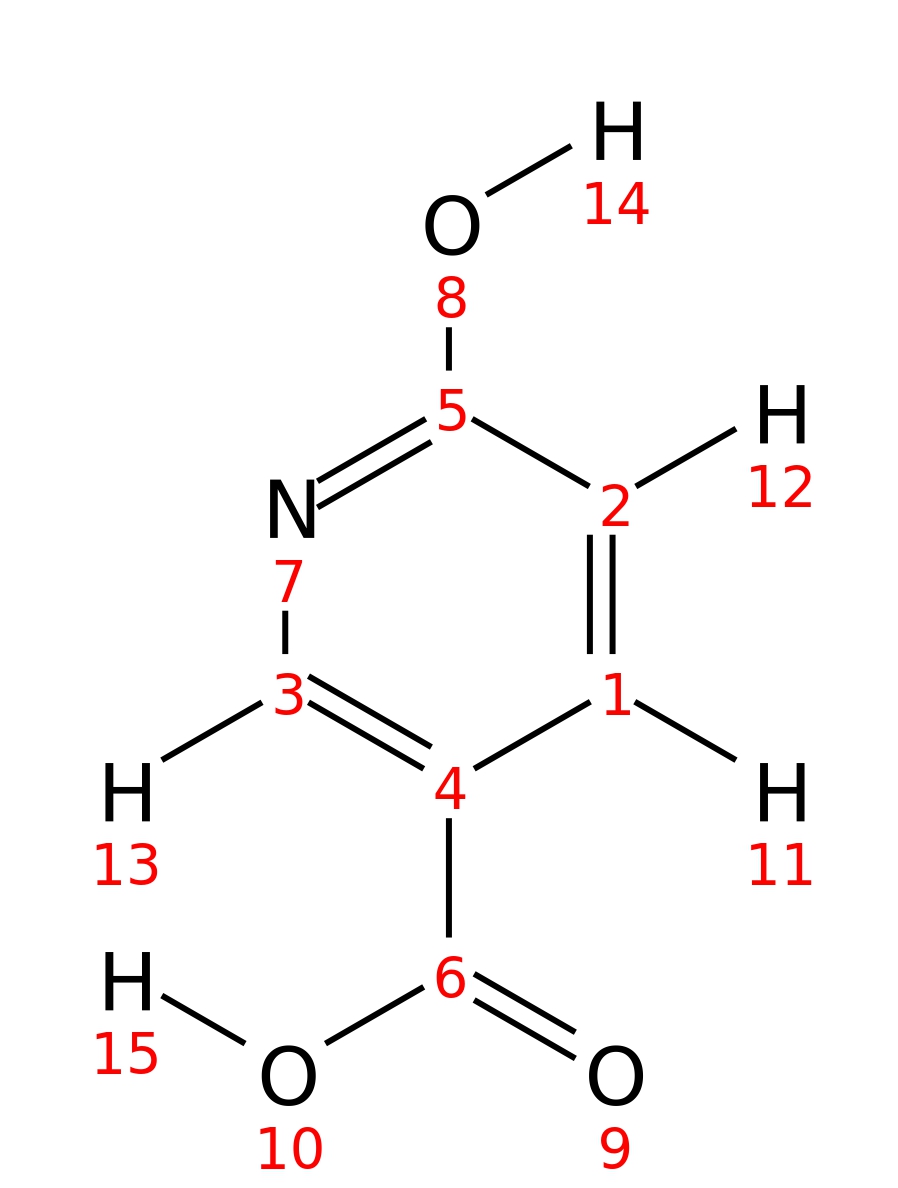
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.00777 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H5NO3/c8-5-2-1-4(3-7-5)6(9)10/h1-3H,(H,7,8)(H,9,10) | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 6-Hydroxynicotinic acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 11 | 12 | 13 | |
|---|---|---|---|
| 11 | 8.084 | 9.285 | 2.144 |
| 12 | 0 | 6.604 | 0 |
| 13 | 0 | 0 | 8.069 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 6.59488 | 0.483915 | standard |
| 6.61295 | 0.483915 | standard |
| 8.07106 | 1.00002 | standard |
| 8.09118 | 0.362721 | standard |
| 8.09481 | 0.292974 | standard |
| -0.995987 | 6.5125e-05 | standard |
| 6.47382 | 0.476823 | standard |
| 6.57719 | 0.141367 | standard |
| 6.61825 | 0.150598 | standard |
| 6.71625 | 0.652528 | standard |
| 8.00259 | 0.829619 | standard |
| 8.04707 | 1.0 | standard |
| 8.08343 | 0.859023 | standard |
| 8.18644 | 0.504352 | standard |
| -0.970695 | 2.8125e-05 | standard |
| 6.51914 | 0.496398 | standard |
| 6.58297 | 0.140815 | standard |
| 6.61924 | 0.151778 | standard |
| 6.68088 | 0.615963 | standard |
| 8.05425 | 1.0 | standard |
| 8.07802 | 0.855374 | standard |
| 8.15024 | 0.510412 | standard |
| -0.997574 | 1.6125e-05 | standard |
| 6.54113 | 0.493289 | standard |
| 6.58436 | 0.139612 | standard |
| 6.62103 | 0.150157 | standard |
| 6.66241 | 0.579874 | standard |
| 8.05745 | 1.0 | standard |
| 8.13288 | 0.496928 | standard |
| -0.959983 | 1.2125e-05 | standard |
| 6.54827 | 0.488008 | standard |
| 6.58439 | 0.139835 | standard |
| 6.65619 | 0.562561 | standard |
| 8.05852 | 1.0 | standard |
| 8.12718 | 0.486546 | standard |
| -0.991722 | 1.0125e-05 | standard |
| 6.55404 | 0.478613 | standard |
| 6.6511 | 0.548437 | standard |
| 8.0593 | 1.0 | standard |
| 8.12266 | 0.47768 | standard |
| -0.85058 | 2.125e-06 | standard |
| 6.57936 | 0.349243 | standard |
| 6.62792 | 0.373777 | standard |
| 8.06484 | 1.0 | standard |
| 8.10274 | 0.341328 | standard |
| 8.11107 | 0.245011 | standard |
| -0.714397 | 1.125e-06 | standard |
| 6.58858 | 0.301556 | standard |
| 6.61909 | 0.310183 | standard |
| 8.0688 | 1.0 | standard |
| 8.09627 | 0.293283 | standard |
| 8.10222 | 0.221336 | standard |
| 6.59286 | 0.355812 | standard |
| 6.61498 | 0.355812 | standard |
| 8.07073 | 1.00021 | standard |
| 8.09308 | 0.29179 | standard |
| 8.09762 | 0.228935 | standard |
| 6.59494 | 0.486339 | standard |
| 6.61295 | 0.484078 | standard |
| 8.07106 | 1.00002 | standard |
| 8.09118 | 0.362759 | standard |
| 8.09481 | 0.292994 | standard |
| 6.59633 | 0.611419 | standard |
| 6.6115 | 0.611407 | standard |
| 8.0705 | 1.00004 | standard |
| 8.07426 | 0.682776 | standard |
| 8.08987 | 0.439311 | standard |
| 8.09293 | 0.362225 | standard |
| 6.59742 | 0.670349 | standard |
| 6.61051 | 0.670349 | standard |
| 8.06782 | 0.736665 | standard |
| 8.0703 | 1.00002 | standard |
| 8.07573 | 0.6038 | standard |
| 8.08896 | 0.470185 | standard |
| 8.09162 | 0.394234 | standard |
| 6.59782 | 0.687386 | standard |
| 6.61006 | 0.685376 | standard |
| 8.06784 | 0.756474 | standard |
| 8.07025 | 1.00052 | standard |
| 8.07627 | 0.584075 | standard |
| 8.08856 | 0.478747 | standard |
| 8.09105 | 0.404268 | standard |
| 6.59822 | 0.699943 | standard |
| 6.60972 | 0.699943 | standard |
| 8.0679 | 0.776673 | standard |
| 8.07014 | 1.00076 | standard |
| 8.07669 | 0.570246 | standard |
| 8.08826 | 0.485287 | standard |
| 8.09057 | 0.413232 | standard |
| 6.59886 | 0.720556 | standard |
| 6.60912 | 0.719526 | standard |
| 8.06803 | 0.805684 | standard |
| 8.07004 | 1.00097 | standard |
| 8.0775 | 0.552479 | standard |
| 8.07948 | 0.418199 | standard |
| 8.08775 | 0.494359 | standard |
| 8.08981 | 0.426135 | standard |
| 6.59911 | 0.723539 | standard |
| 6.60882 | 0.723539 | standard |
| 8.06811 | 0.816987 | standard |
| 8.06999 | 1.00001 | standard |
| 8.0778 | 0.543118 | standard |
| 8.07974 | 0.419025 | standard |
| 8.08755 | 0.493712 | standard |
| 8.08953 | 0.427093 | standard |
| 6.59936 | 0.733781 | standard |
| 6.60858 | 0.733781 | standard |
| 8.06813 | 0.829019 | standard |
| 8.06995 | 1.00107 | standard |
| 8.07811 | 0.544883 | standard |
| 8.07983 | 0.433453 | standard |
| 8.08735 | 0.503007 | standard |
| 8.08912 | 0.440126 | standard |
| 6.59975 | 0.743399 | standard |
| 6.60818 | 0.743399 | standard |
| 8.0682 | 0.84668 | standard |
| 8.06985 | 1.0011 | standard |
| 8.07861 | 0.538612 | standard |
| 8.08018 | 0.442318 | standard |
| 8.08694 | 0.500776 | standard |
| 8.08868 | 0.438844 | standard |
| 6.60045 | 0.749409 | standard |
| 6.60754 | 0.747149 | standard |
| 8.06837 | 0.87344 | standard |
| 8.0697 | 1.00011 | standard |
| 8.07929 | 0.518164 | standard |
| 8.08076 | 0.439549 | standard |
| 8.08643 | 0.499053 | standard |
| 8.08791 | 0.443812 | standard |