3-5-Dichlorocatechol
Simulation outputs:
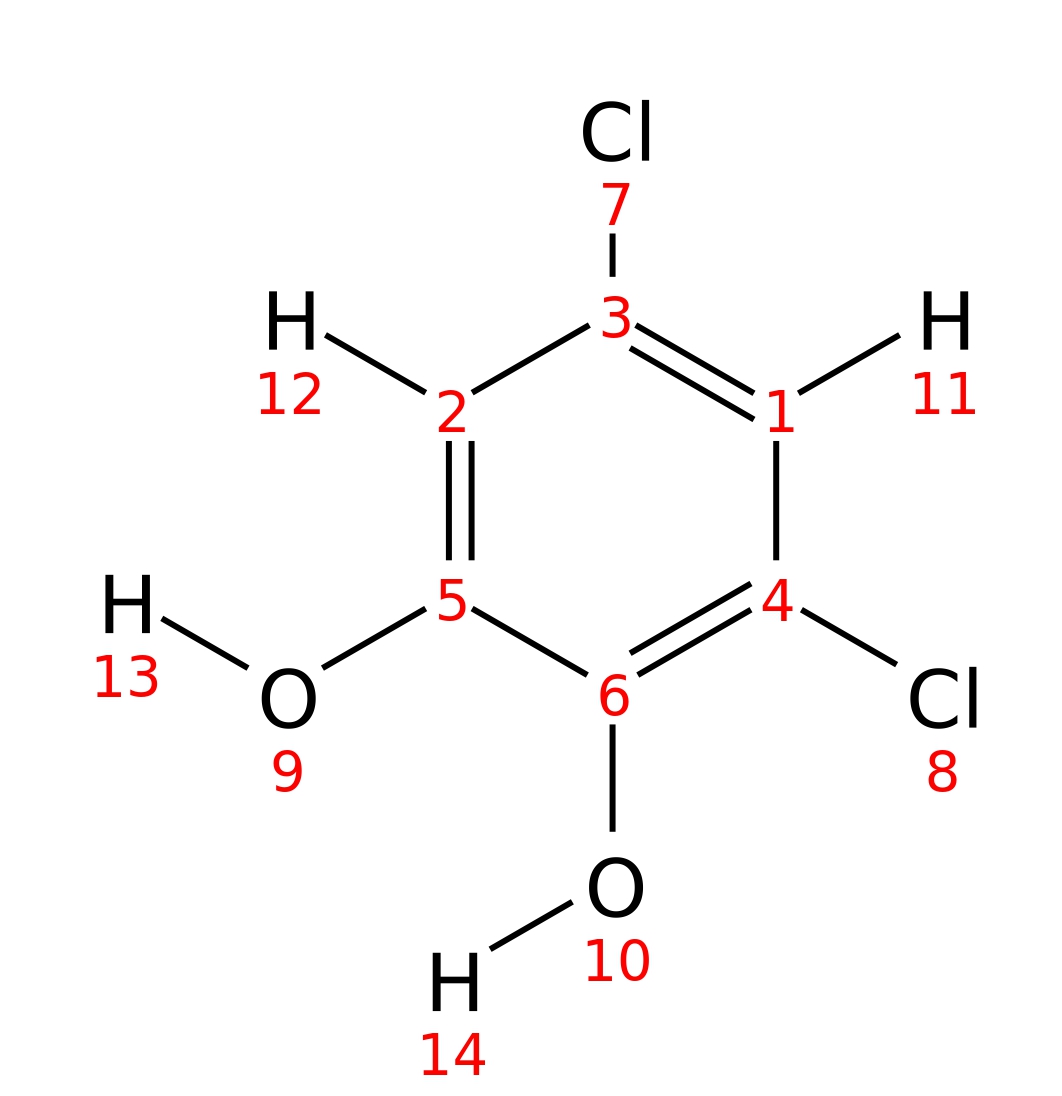
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.00662 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H4Cl2O2/c7-3-1-4(8)6(10)5(9)2-3/h1-2,9-10H | |
| Note 1 | 11?12 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3,5-dichlorocatechol | Solute | SaturatedmM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 11 | 12 | |
|---|---|---|
| 11 | 6.908 | 2.469 |
| 12 | 0 | 6.805 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 6.80221 | 0.931187 | standard |
| 6.80696 | 1.00001 | standard |
| 6.90554 | 1.00001 | standard |
| 6.91029 | 0.931187 | standard |
| -0.95175 | 9.125e-06 | standard |
| 6.77184 | 0.431028 | standard |
| 6.82815 | 1.0 | standard |
| 6.88435 | 1.0 | standard |
| 6.94066 | 0.431028 | standard |
| -0.95304 | 5.125e-06 | standard |
| 6.78234 | 0.562677 | standard |
| 6.82108 | 1.00001 | standard |
| 6.89141 | 1.00001 | standard |
| 6.93016 | 0.562677 | standard |
| -0.898685 | 3.125e-06 | standard |
| 6.78822 | 0.646577 | standard |
| 6.81748 | 1.0 | standard |
| 6.89502 | 1.0 | standard |
| 6.92428 | 0.646577 | standard |
| -0.732251 | 2.125e-06 | standard |
| 6.79015 | 0.677095 | standard |
| 6.81628 | 0.999531 | standard |
| 6.89632 | 1.0 | standard |
| 6.92233 | 0.677981 | standard |
| -0.835305 | 2.125e-06 | standard |
| 6.79168 | 0.703825 | standard |
| 6.81519 | 1.0 | standard |
| 6.89731 | 1.0 | standard |
| 6.92082 | 0.703825 | standard |
| -0.951353 | 1.125e-06 | standard |
| 6.79836 | 0.837183 | standard |
| 6.81023 | 1.0 | standard |
| 6.90227 | 1.0 | standard |
| 6.91414 | 0.837183 | standard |
| 6.80051 | 0.88845 | standard |
| 6.80839 | 1.00017 | standard |
| 6.9041 | 1.00017 | standard |
| 6.91198 | 0.88845 | standard |
| 6.80154 | 0.911547 | standard |
| 6.80754 | 0.996872 | standard |
| 6.90506 | 1.00004 | standard |
| 6.91095 | 0.915152 | standard |
| 6.80221 | 0.931197 | standard |
| 6.80696 | 1.00001 | standard |
| 6.90554 | 1.00001 | standard |
| 6.91029 | 0.931197 | standard |
| 6.80258 | 0.942198 | standard |
| 6.80658 | 1.00014 | standard |
| 6.90592 | 1.00014 | standard |
| 6.90991 | 0.942198 | standard |
| 6.80286 | 0.950513 | standard |
| 6.80629 | 1.0004 | standard |
| 6.90621 | 1.0004 | standard |
| 6.90963 | 0.950513 | standard |
| 6.80306 | 0.954292 | standard |
| 6.80619 | 1.00049 | standard |
| 6.9063 | 0.994309 | standard |
| 6.90954 | 0.947659 | standard |
| 6.80315 | 0.957271 | standard |
| 6.8061 | 1.0007 | standard |
| 6.90639 | 0.994134 | standard |
| 6.90945 | 0.950252 | standard |
| 6.80324 | 0.962294 | standard |
| 6.80591 | 1.00136 | standard |
| 6.90659 | 1.00136 | standard |
| 6.90925 | 0.962294 | standard |
| 6.80334 | 0.964833 | standard |
| 6.80581 | 1.00148 | standard |
| 6.90669 | 1.00148 | standard |
| 6.90915 | 0.964833 | standard |
| 6.80344 | 0.966775 | standard |
| 6.80581 | 1.00201 | standard |
| 6.90668 | 1.00201 | standard |
| 6.90906 | 0.966775 | standard |
| 6.80353 | 0.960921 | standard |
| 6.80572 | 0.992793 | standard |
| 6.90688 | 1.00237 | standard |
| 6.90896 | 0.970787 | standard |
| 6.80372 | 1.00235 | standard |
| 6.80552 | 1.00236 | standard |
| 6.90697 | 1.00236 | standard |
| 6.90878 | 1.00235 | standard |