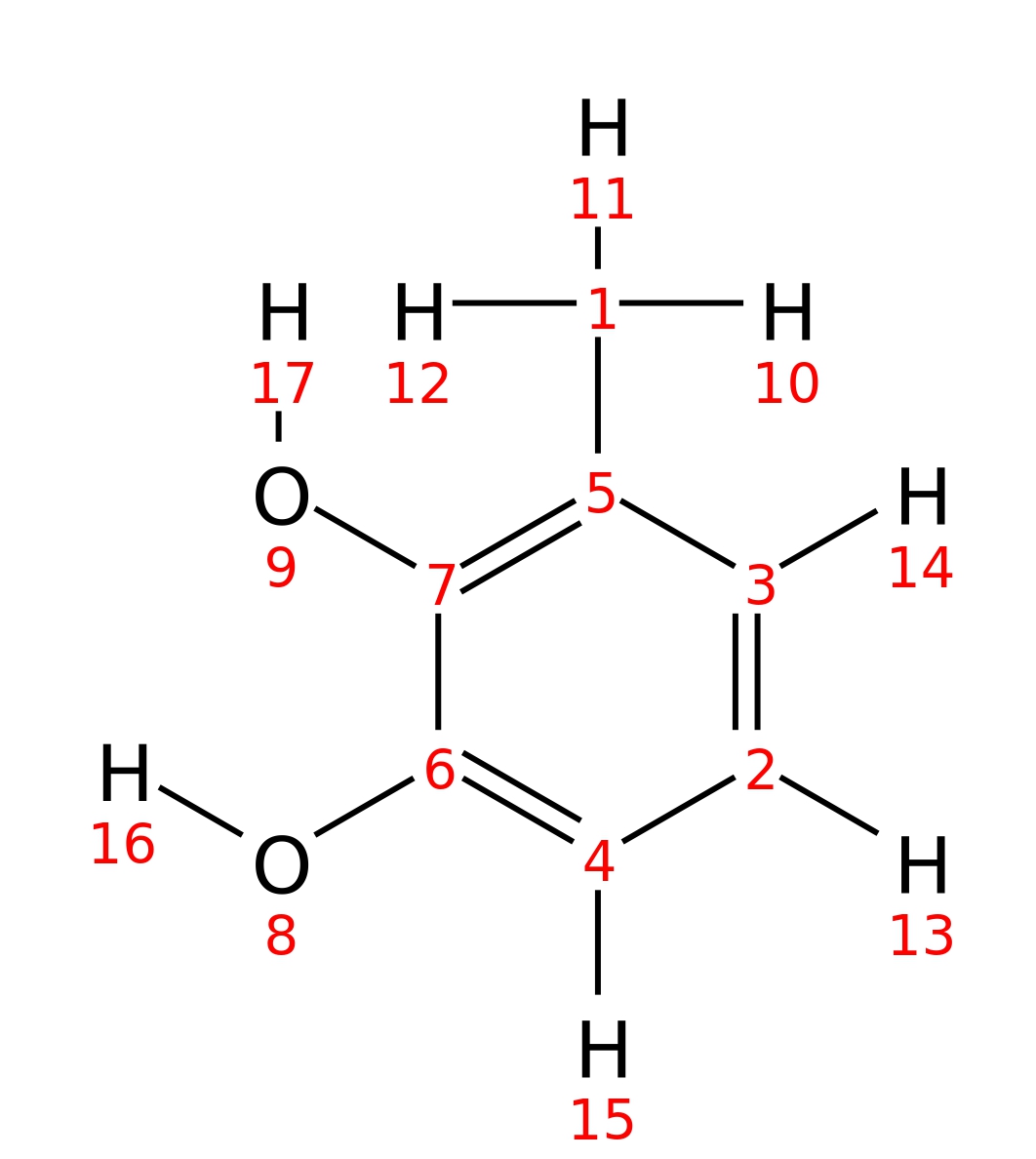
3-Methylcatechol
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.00662 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H8O2/c1-5-3-2-4-6(8)7(5)9/h2-4,8-9H,1H3 | |
| Note 1 | 3 overlaping coupled protons | |
| Note 2 | 14?15 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-Methylcatechol | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 10 | 11 | 12 | 14 | 13 | 15 | |
|---|---|---|---|---|---|---|
| 10 | 2.216 | -14.9 | -14.9 | 0 | 0 | 0 |
| 11 | 0 | 2.216 | -14.9 | 0 | 0 | 0 |
| 12 | 0 | 0 | 2.216 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 6.794 | 7.427 | 0.983 |
| 13 | 0 | 0 | 0 | 0 | 6.784 | 8.061 |
| 15 | 0 | 0 | 0 | 0 | 0 | 6.772 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.21631 | 1.0 | standard |
| 6.75793 | 0.0184292 | standard |
| 6.77133 | 0.141998 | standard |
| 6.77819 | 0.243407 | standard |
| 6.78382 | 0.377675 | standard |
| 6.78917 | 0.246 | standard |
| 6.79695 | 0.160799 | standard |
| 2.21631 | 1.0 | standard |
| 6.78377 | 0.994936 | standard |
| 2.21631 | 1.0 | standard |
| 6.78375 | 0.988581 | standard |
| 2.21631 | 1.0 | standard |
| 6.78374 | 0.977999 | standard |
| 2.21631 | 1.0 | standard |
| 6.78376 | 0.972429 | standard |
| 2.21631 | 1.0 | standard |
| 6.78374 | 0.964643 | standard |
| 2.21631 | 1.0 | standard |
| 6.78379 | 0.815245 | standard |
| 6.80832 | 0.080202 | standard |
| 2.21631 | 1.0 | standard |
| 6.76555 | 0.0988195 | standard |
| 6.7838 | 0.613341 | standard |
| 6.80307 | 0.103466 | standard |
| 2.21631 | 1.0 | standard |
| 6.76912 | 0.120677 | standard |
| 6.78384 | 0.469369 | standard |
| 6.79939 | 0.132514 | standard |
| 2.21631 | 1.0 | standard |
| 6.75793 | 0.0184282 | standard |
| 6.77133 | 0.141995 | standard |
| 6.77819 | 0.243339 | standard |
| 6.78382 | 0.377561 | standard |
| 6.78917 | 0.245906 | standard |
| 6.79695 | 0.160419 | standard |
| 2.21631 | 1.0 | standard |
| 6.76094 | 0.0230293 | standard |
| 6.76685 | 0.0551206 | standard |
| 6.7732 | 0.157154 | standard |
| 6.77739 | 0.215169 | standard |
| 6.78383 | 0.313564 | standard |
| 6.78986 | 0.21495 | standard |
| 6.7953 | 0.18416 | standard |
| 2.21631 | 1.0 | standard |
| 6.76302 | 0.028257 | standard |
| 6.76725 | 0.0537988 | standard |
| 6.77673 | 0.213185 | standard |
| 6.78395 | 0.266756 | standard |
| 6.79045 | 0.203637 | standard |
| 6.79419 | 0.198327 | standard |
| 6.79938 | 0.058366 | standard |
| 2.21631 | 1.0 | standard |
| 6.76388 | 0.0310377 | standard |
| 6.7676 | 0.0547458 | standard |
| 6.77641 | 0.223602 | standard |
| 6.78397 | 0.250336 | standard |
| 6.79071 | 0.203835 | standard |
| 6.7937 | 0.203816 | standard |
| 6.79909 | 0.0581984 | standard |
| 2.21631 | 1.0 | standard |
| 6.76456 | 0.0339215 | standard |
| 6.76774 | 0.0550799 | standard |
| 6.77348 | 0.147541 | standard |
| 6.7763 | 0.23848 | standard |
| 6.78397 | 0.234947 | standard |
| 6.79086 | 0.206182 | standard |
| 6.79311 | 0.199017 | standard |
| 6.79886 | 0.0582698 | standard |
| 6.802 | 0.0298021 | standard |
| 2.21631 | 1.0 | standard |
| 6.76574 | 0.039828 | standard |
| 6.76819 | 0.0575436 | standard |
| 6.77377 | 0.150429 | standard |
| 6.77627 | 0.248402 | standard |
| 6.784 | 0.215798 | standard |
| 6.79139 | 0.23301 | standard |
| 6.79845 | 0.0592912 | standard |
| 6.80091 | 0.0353151 | standard |
| 2.21631 | 1.0 | standard |
| 6.76625 | 0.0429732 | standard |
| 6.76836 | 0.0590189 | standard |
| 6.77383 | 0.149307 | standard |
| 6.77629 | 0.227497 | standard |
| 6.78401 | 0.207199 | standard |
| 6.79148 | 0.253042 | standard |
| 6.79828 | 0.0600772 | standard |
| 6.8004 | 0.0382472 | standard |
| 2.21631 | 1.0 | standard |
| 6.76664 | 0.0456708 | standard |
| 6.76855 | 0.060536 | standard |
| 6.77393 | 0.152369 | standard |
| 6.77601 | 0.204484 | standard |
| 6.78402 | 0.200503 | standard |
| 6.79141 | 0.260725 | standard |
| 6.7981 | 0.0610948 | standard |
| 6.80005 | 0.0408446 | standard |
| 2.21631 | 1.0 | standard |
| 6.76732 | 0.0505479 | standard |
| 6.76892 | 0.0635205 | standard |
| 6.774 | 0.152714 | standard |
| 6.77567 | 0.180755 | standard |
| 6.7776 | 0.126664 | standard |
| 6.78413 | 0.188969 | standard |
| 6.79133 | 0.238116 | standard |
| 6.79313 | 0.168745 | standard |
| 6.79784 | 0.0635494 | standard |
| 2.21631 | 1.0 | standard |
| 6.76837 | 0.0605801 | standard |
| 6.76938 | 0.069884 | standard |
| 6.77412 | 0.157232 | standard |
| 6.7752 | 0.169185 | standard |
| 6.77866 | 0.102271 | standard |
| 6.78415 | 0.178711 | standard |
| 6.79011 | 0.127998 | standard |
| 6.79195 | 0.182513 | standard |
| 6.79306 | 0.166589 | standard |
| 6.7974 | 0.0704057 | standard |