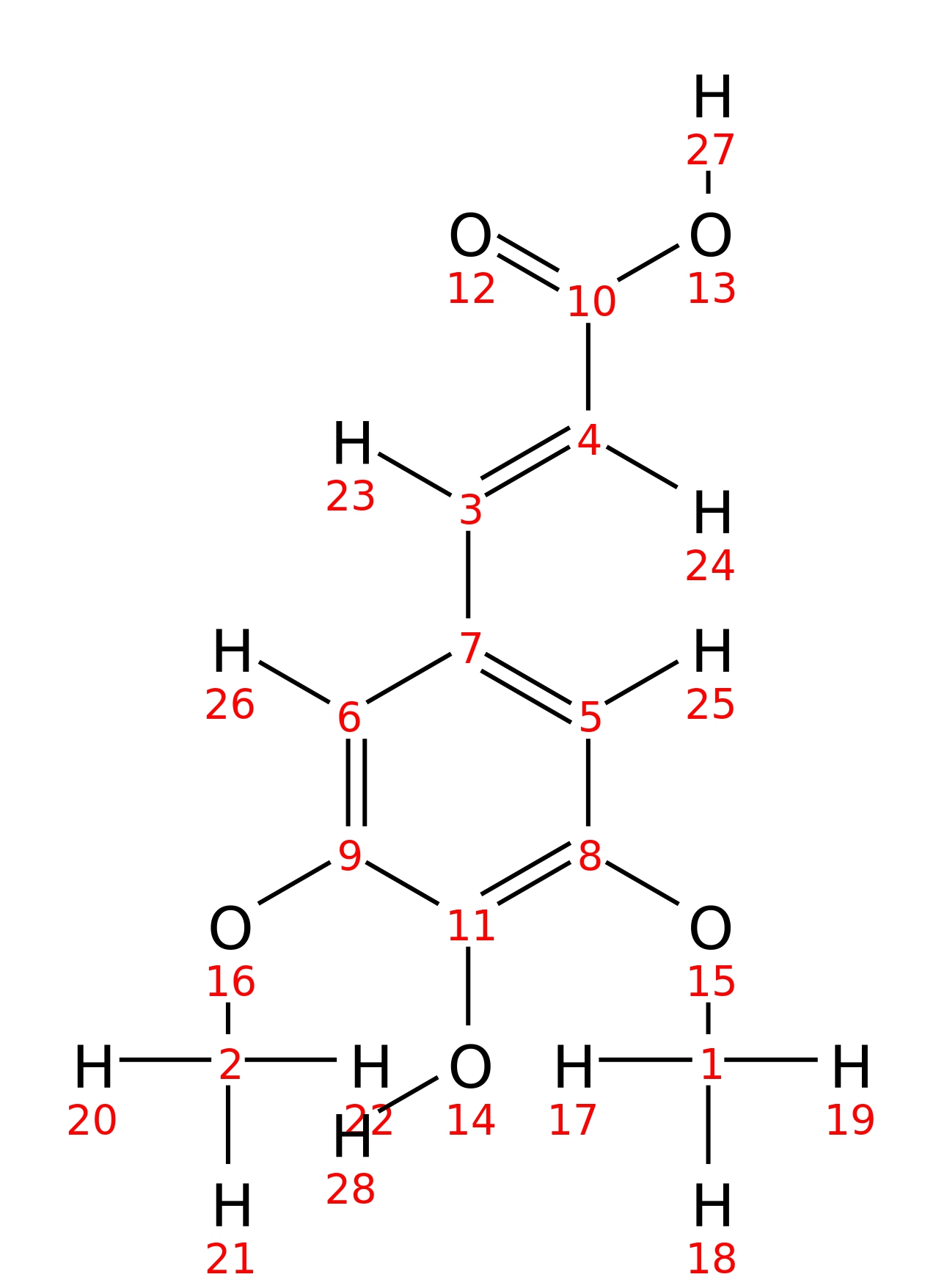
Sinapic-acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01620 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H12O5/c1-15-8-5-7(3-4-10(12)13)6-9(16-2)11(8)14/h3-6,14H,1-2H3,(H,12,13)/b4-3+ | |
| Note 1 | 23?24 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| sinapic acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 17 | 18 | 19 | 25 | 26 | 20 | 21 | 22 | 23 | 24 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 17 | 3.814 | -11.5 | -11.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 3.814 | -11.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 3.814 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 6.71 | 2.554 | 0 | 0 | 0 | 0.452 | 0 |
| 26 | 0 | 0 | 0 | 0 | 6.709 | 0 | 0 | 0 | 1.093 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 3.814 | -11.5 | -11.5 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 3.814 | -11.5 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.814 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.192 | 16.423 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.301 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.8142 | 1.0 | standard |
| 6.28444 | 0.080746 | standard |
| 6.31727 | 0.086876 | standard |
| 6.70957 | 0.286834 | standard |
| 7.17637 | 0.0701203 | standard |
| 7.20925 | 0.064508 | standard |
| 3.8142 | 1.0 | standard |
| 6.0511 | 0.0494563 | standard |
| 6.46131 | 0.12225 | standard |
| 6.7101 | 0.295584 | standard |
| 7.03181 | 0.099332 | standard |
| 7.44269 | 0.0406341 | standard |
| 3.8142 | 1.0 | standard |
| 6.14384 | 0.0599464 | standard |
| 6.41743 | 0.10943 | standard |
| 6.70979 | 0.292319 | standard |
| 7.07607 | 0.0888161 | standard |
| 7.34985 | 0.0483382 | standard |
| 3.8142 | 1.0 | standard |
| 6.18692 | 0.0656131 | standard |
| 6.39213 | 0.103299 | standard |
| 6.70966 | 0.291439 | standard |
| 7.10147 | 0.0832972 | standard |
| 7.30683 | 0.052872 | standard |
| 3.8142 | 1.0 | standard |
| 6.20073 | 0.067655 | standard |
| 6.38313 | 0.101268 | standard |
| 6.70963 | 0.290765 | standard |
| 7.11054 | 0.0818151 | standard |
| 7.29297 | 0.054013 | standard |
| 3.8142 | 1.0 | standard |
| 6.21156 | 0.0691106 | standard |
| 6.37579 | 0.099368 | standard |
| 6.70959 | 0.290849 | standard |
| 7.11791 | 0.080325 | standard |
| 7.2822 | 0.0557258 | standard |
| 3.8142 | 1.0 | standard |
| 6.25825 | 0.076386 | standard |
| 6.34038 | 0.091703 | standard |
| 6.70956 | 0.28991 | standard |
| 7.15336 | 0.0741481 | standard |
| 7.2355 | 0.0615578 | standard |
| 3.8142 | 1.0 | standard |
| 6.27293 | 0.078919 | standard |
| 6.32768 | 0.08915 | standard |
| 6.70957 | 0.289762 | standard |
| 7.16595 | 0.0718216 | standard |
| 7.22069 | 0.0635955 | standard |
| 3.8142 | 1.0 | standard |
| 6.28017 | 0.079993 | standard |
| 6.32124 | 0.087652 | standard |
| 6.70959 | 0.28912 | standard |
| 7.17244 | 0.0705657 | standard |
| 7.21352 | 0.064889 | standard |
| 3.8142 | 1.0 | standard |
| 6.28444 | 0.080747 | standard |
| 6.31727 | 0.086875 | standard |
| 6.70957 | 0.286808 | standard |
| 7.17637 | 0.0701133 | standard |
| 7.20926 | 0.0651089 | standard |
| 3.8142 | 1.0 | standard |
| 6.28732 | 0.081603 | standard |
| 6.31469 | 0.08623 | standard |
| 6.70954 | 0.290919 | standard |
| 7.17906 | 0.0694713 | standard |
| 7.20644 | 0.0659073 | standard |
| 3.8142 | 1.0 | standard |
| 6.2893 | 0.082179 | standard |
| 6.31281 | 0.085065 | standard |
| 6.70959 | 0.286302 | standard |
| 7.18093 | 0.0676811 | standard |
| 7.20439 | 0.066432 | standard |
| 3.8142 | 1.0 | standard |
| 6.29009 | 0.082788 | standard |
| 6.31201 | 0.084832 | standard |
| 6.70954 | 0.287225 | standard |
| 7.18173 | 0.0679506 | standard |
| 7.2036 | 0.067439 | standard |
| 3.8142 | 1.0 | standard |
| 6.29079 | 0.082856 | standard |
| 6.31132 | 0.084772 | standard |
| 6.70958 | 0.285577 | standard |
| 7.18237 | 0.0674 | standard |
| 7.2029 | 0.066227 | standard |
| 3.8142 | 1.0 | standard |
| 6.29198 | 0.082972 | standard |
| 6.31023 | 0.084676 | standard |
| 6.70954 | 0.288713 | standard |
| 7.18345 | 0.0681983 | standard |
| 7.20171 | 0.066001 | standard |
| 3.8142 | 1.0 | standard |
| 6.29247 | 0.08383 | standard |
| 6.30973 | 0.08383 | standard |
| 6.70957 | 0.288571 | standard |
| 7.18396 | 0.067739 | standard |
| 7.20128 | 0.068029 | standard |
| 3.8142 | 1.0 | standard |
| 6.29287 | 0.083836 | standard |
| 6.30933 | 0.083836 | standard |
| 6.70958 | 0.284543 | standard |
| 7.18442 | 0.0670369 | standard |
| 7.20076 | 0.0670369 | standard |
| 3.8142 | 1.0 | standard |
| 6.29366 | 0.08386 | standard |
| 6.30854 | 0.083861 | standard |
| 6.70956 | 0.287023 | standard |
| 7.18509 | 0.0677938 | standard |
| 7.20003 | 0.067405 | standard |
| 3.8142 | 1.0 | standard |
| 6.29485 | 0.083915 | standard |
| 6.30745 | 0.083915 | standard |
| 6.70958 | 0.284498 | standard |
| 7.18624 | 0.067031 | standard |
| 7.1989 | 0.0675731 | standard |