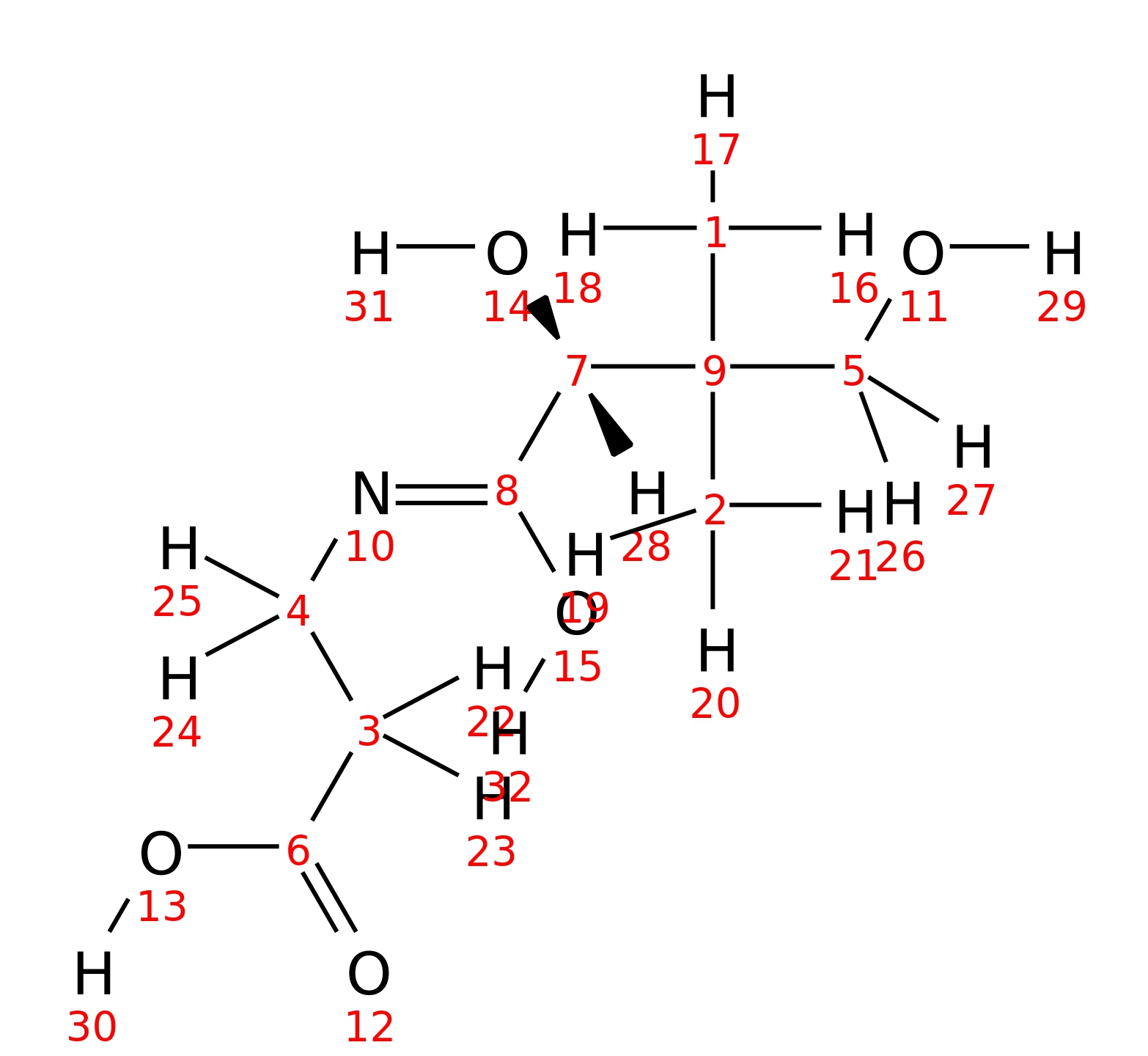
Pantothenate
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.01005 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H17NO5/c1-9(2,5-11)7(14)8(15)10-4-3-6(12)13/h7,11,14H,3-5H2,1-2H3,(H,10,15)(H,12,13)/t7-/m0/s1 | |
| Note 1 | 22,23?24,25 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Pantothenate | Solute | Saturated1 |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 16 | 17 | 18 | 19 | 20 | 21 | 26 | 27 | 28 | 24 | 25 | 22 | 23 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 0.919 | -12.5 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0.919 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0.919 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0.883 | -12.5 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0.883 | -12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0.883 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 3.385 | -11.164 | 0 | 0 | 0 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.502 | 0 | 0 | 0 | 0 | 0 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.972 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.429 | -12.4 | 5.325 | 10.375 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.429 | 3.386 | 8.038 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.406 | -12.4 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.406 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 0.882652 | 0.998341 | standard |
| 0.919548 | 1.0 | standard |
| 2.35661 | 0.00471375 | standard |
| 2.38899 | 0.129604 | standard |
| 2.40563 | 0.312486 | standard |
| 2.42255 | 0.145923 | standard |
| 2.45442 | 0.00521556 | standard |
| 3.36936 | 0.122923 | standard |
| 3.39733 | 0.205973 | standard |
| 3.41215 | 0.172945 | standard |
| 3.42908 | 0.263255 | standard |
| 3.446 | 0.158848 | standard |
| 3.46071 | 0.0149561 | standard |
| 3.48947 | 0.194684 | standard |
| 3.51743 | 0.122083 | standard |
| 3.97171 | 0.311627 | standard |
| 0.887645 | 1.0 | standard |
| 0.914554 | 0.99993 | standard |
| 1.89123 | 0.00358913 | standard |
| 2.19742 | 0.0515623 | standard |
| 2.25098 | 0.0607851 | standard |
| 2.39683 | 0.174007 | standard |
| 2.5552 | 0.163391 | standard |
| 2.8575 | 0.00987013 | standard |
| 2.99725 | 0.00710112 | standard |
| 3.15179 | 0.0291411 | standard |
| 3.27989 | 0.160026 | standard |
| 3.44667 | 0.541401 | standard |
| 3.59267 | 0.0793471 | standard |
| 3.62253 | 0.0707521 | standard |
| 3.73203 | 0.0232081 | standard |
| 3.9717 | 0.236206 | standard |
| 0.883802 | 0.999889 | standard |
| 0.918399 | 1.0 | standard |
| 2.06788 | 0.00361213 | standard |
| 2.27453 | 0.071082 | standard |
| 2.30021 | 0.077913 | standard |
| 2.39878 | 0.230019 | standard |
| 2.51035 | 0.168153 | standard |
| 2.71636 | 0.00722112 | standard |
| 3.24056 | 0.0386941 | standard |
| 3.3245 | 0.174319 | standard |
| 3.43173 | 0.498119 | standard |
| 3.54268 | 0.107809 | standard |
| 3.64544 | 0.0332401 | standard |
| 3.97171 | 0.268319 | standard |
| 0.883055 | 0.999363 | standard |
| 0.919146 | 1.0 | standard |
| 2.15427 | 0.00380912 | standard |
| 2.31103 | 0.08453 | standard |
| 2.32556 | 0.0899126 | standard |
| 2.40166 | 0.262041 | standard |
| 2.48587 | 0.166818 | standard |
| 2.64207 | 0.00650312 | standard |
| 3.28261 | 0.0508004 | standard |
| 3.34886 | 0.180601 | standard |
| 3.42536 | 0.462179 | standard |
| 3.46327 | 0.329583 | standard |
| 3.51517 | 0.131728 | standard |
| 3.60338 | 0.0442674 | standard |
| 3.97171 | 0.284546 | standard |
| 0.882911 | 1.00001 | standard |
| 0.919288 | 0.998562 | standard |
| 2.18284 | 0.00391113 | standard |
| 2.32279 | 0.0894574 | standard |
| 2.33413 | 0.0941951 | standard |
| 2.40251 | 0.271345 | standard |
| 2.4775 | 0.165434 | standard |
| 2.61663 | 0.006267 | standard |
| 3.29615 | 0.056765 | standard |
| 3.35726 | 0.182738 | standard |
| 3.42341 | 0.438722 | standard |
| 3.466 | 0.309318 | standard |
| 3.50523 | 0.142945 | standard |
| 3.59021 | 0.0489762 | standard |
| 3.97171 | 0.289774 | standard |
| 0.882827 | 0.998166 | standard |
| 0.919374 | 1.00001 | standard |
| 2.20565 | 0.00398112 | standard |
| 2.33271 | 0.0941261 | standard |
| 2.34088 | 0.098198 | standard |
| 2.40316 | 0.278737 | standard |
| 2.47061 | 0.164132 | standard |
| 2.59634 | 0.00612325 | standard |
| 3.30686 | 0.062314 | standard |
| 3.3641 | 0.184044 | standard |
| 3.42167 | 0.409921 | standard |
| 3.46816 | 0.298063 | standard |
| 3.49692 | 0.155896 | standard |
| 3.57978 | 0.0538479 | standard |
| 3.97171 | 0.293688 | standard |
| 0.882664 | 1.00001 | standard |
| 0.919537 | 0.999956 | standard |
| 2.30672 | 0.00435013 | standard |
| 2.37249 | 0.118137 | standard |
| 2.40514 | 0.304224 | standard |
| 2.43891 | 0.153229 | standard |
| 2.50238 | 0.00554004 | standard |
| 3.35075 | 0.094688 | standard |
| 3.3964 | 0.217829 | standard |
| 3.40632 | 0.271237 | standard |
| 3.42943 | 0.273628 | standard |
| 3.46365 | 0.168851 | standard |
| 3.48014 | 0.240325 | standard |
| 3.53607 | 0.0919271 | standard |
| 3.97171 | 0.307984 | standard |
| 0.882654 | 0.99927 | standard |
| 0.919546 | 1.0 | standard |
| 2.34005 | 0.0044425 | standard |
| 2.38342 | 0.126134 | standard |
| 2.40553 | 0.310161 | standard |
| 2.42804 | 0.149202 | standard |
| 2.47056 | 0.00543523 | standard |
| 3.36351 | 0.112689 | standard |
| 3.40109 | 0.25429 | standard |
| 3.4059 | 0.22729 | standard |
| 3.42919 | 0.265716 | standard |
| 3.45174 | 0.15675 | standard |
| 3.48609 | 0.207472 | standard |
| 3.52328 | 0.111339 | standard |
| 3.97171 | 0.31098 | standard |
| 0.882652 | 0.999106 | standard |
| 0.919548 | 1.0 | standard |
| 2.35652 | 0.00470656 | standard |
| 2.38899 | 0.129673 | standard |
| 2.40563 | 0.312612 | standard |
| 2.42255 | 0.145996 | standard |
| 2.45442 | 0.00521856 | standard |
| 3.36936 | 0.122961 | standard |
| 3.39733 | 0.206068 | standard |
| 3.41215 | 0.173022 | standard |
| 3.42908 | 0.263387 | standard |
| 3.446 | 0.158916 | standard |
| 3.46071 | 0.0149681 | standard |
| 3.48947 | 0.194771 | standard |
| 3.51743 | 0.12212 | standard |
| 3.97171 | 0.311745 | standard |
| 0.882652 | 0.998665 | standard |
| 0.919549 | 1.0 | standard |
| 2.3665 | 0.0049262 | standard |
| 2.39235 | 0.13326 | standard |
| 2.4057 | 0.313848 | standard |
| 2.41918 | 0.144531 | standard |
| 2.44472 | 0.00508117 | standard |
| 3.37273 | 0.129487 | standard |
| 3.39514 | 0.190149 | standard |
| 3.41549 | 0.167743 | standard |
| 3.42902 | 0.260285 | standard |
| 3.44253 | 0.160034 | standard |
| 3.45427 | 0.0137391 | standard |
| 3.49165 | 0.187371 | standard |
| 3.51406 | 0.128902 | standard |
| 3.97171 | 0.31207 | standard |
| 0.882652 | 1.0 | standard |
| 0.919549 | 0.999617 | standard |
| 2.37303 | 0.004932 | standard |
| 2.39454 | 0.133507 | standard |
| 2.40575 | 0.313661 | standard |
| 2.41699 | 0.141513 | standard |
| 2.43829 | 0.004905 | standard |
| 3.37491 | 0.134175 | standard |
| 3.39355 | 0.184127 | standard |
| 3.40779 | 0.0159923 | standard |
| 3.41767 | 0.165692 | standard |
| 3.42903 | 0.26082 | standard |
| 3.44025 | 0.161345 | standard |
| 3.45003 | 0.0133275 | standard |
| 3.49324 | 0.182714 | standard |
| 3.51188 | 0.133744 | standard |
| 3.97171 | 0.3127 | standard |
| 0.882652 | 0.998705 | standard |
| 0.919549 | 1.0 | standard |
| 2.37777 | 0.0049382 | standard |
| 2.39613 | 0.135253 | standard |
| 2.40576 | 0.314044 | standard |
| 2.41542 | 0.141406 | standard |
| 2.43364 | 0.00491114 | standard |
| 3.3765 | 0.137406 | standard |
| 3.39246 | 0.180013 | standard |
| 3.41091 | 0.014199 | standard |
| 3.4193 | 0.16442 | standard |
| 3.42897 | 0.262324 | standard |
| 3.43867 | 0.161565 | standard |
| 3.44698 | 0.0132897 | standard |
| 3.49433 | 0.179132 | standard |
| 3.51029 | 0.137074 | standard |
| 3.97171 | 0.312705 | standard |
| 0.882652 | 0.999641 | standard |
| 0.919549 | 1.0 | standard |
| 2.37965 | 0.00493748 | standard |
| 2.39686 | 0.13241 | standard |
| 2.40576 | 0.313737 | standard |
| 2.41478 | 0.142088 | standard |
| 2.43186 | 0.0048994 | standard |
| 3.37709 | 0.138806 | standard |
| 3.39197 | 0.178542 | standard |
| 3.41214 | 0.0138655 | standard |
| 3.41995 | 0.16382 | standard |
| 3.42896 | 0.261319 | standard |
| 3.43798 | 0.161298 | standard |
| 3.44576 | 0.0131827 | standard |
| 3.49482 | 0.177818 | standard |
| 3.5097 | 0.138511 | standard |
| 3.97171 | 0.312908 | standard |
| 0.882652 | 0.999894 | standard |
| 0.919549 | 1.0 | standard |
| 2.38123 | 0.00491997 | standard |
| 2.3974 | 0.136432 | standard |
| 2.40578 | 0.314628 | standard |
| 2.41418 | 0.14 | standard |
| 2.43018 | 0.00491173 | standard |
| 3.37759 | 0.139985 | standard |
| 3.39157 | 0.177214 | standard |
| 3.41317 | 0.0135805 | standard |
| 3.42049 | 0.164223 | standard |
| 3.42897 | 0.264643 | standard |
| 3.43743 | 0.162322 | standard |
| 3.44474 | 0.0131092 | standard |
| 3.49522 | 0.176606 | standard |
| 3.5092 | 0.139721 | standard |
| 3.97171 | 0.312995 | standard |
| 0.882652 | 0.999484 | standard |
| 0.919549 | 1.0 | standard |
| 2.384 | 0.004927 | standard |
| 2.39835 | 0.13522 | standard |
| 2.40578 | 0.313707 | standard |
| 2.41324 | 0.139249 | standard |
| 2.42751 | 0.00490289 | standard |
| 3.37848 | 0.141926 | standard |
| 3.39087 | 0.174973 | standard |
| 3.4149 | 0.0132621 | standard |
| 3.42143 | 0.163265 | standard |
| 3.42896 | 0.262734 | standard |
| 3.43644 | 0.161814 | standard |
| 3.44298 | 0.0129989 | standard |
| 3.49591 | 0.174526 | standard |
| 3.50831 | 0.141711 | standard |
| 3.97171 | 0.313002 | standard |
| 0.882652 | 1.0 | standard |
| 0.919549 | 0.999776 | standard |
| 2.38519 | 0.0049427 | standard |
| 2.39868 | 0.136711 | standard |
| 2.40579 | 0.3142 | standard |
| 2.4129 | 0.140938 | standard |
| 2.42632 | 0.00492334 | standard |
| 3.37878 | 0.142761 | standard |
| 3.39058 | 0.174065 | standard |
| 3.41569 | 0.0132427 | standard |
| 3.42183 | 0.163143 | standard |
| 3.42893 | 0.264369 | standard |
| 3.4361 | 0.162905 | standard |
| 3.44221 | 0.0130526 | standard |
| 3.49621 | 0.173675 | standard |
| 3.50801 | 0.142566 | standard |
| 3.97171 | 0.313075 | standard |
| 0.882652 | 0.997768 | standard |
| 0.919549 | 1.0 | standard |
| 2.38618 | 0.00492168 | standard |
| 2.39909 | 0.134641 | standard |
| 2.40578 | 0.314934 | standard |
| 2.4125 | 0.139092 | standard |
| 2.42533 | 0.00490217 | standard |
| 3.37917 | 0.143395 | standard |
| 3.39028 | 0.173089 | standard |
| 3.41631 | 0.0131156 | standard |
| 3.42218 | 0.163803 | standard |
| 3.42893 | 0.260057 | standard |
| 3.4357 | 0.162695 | standard |
| 3.44156 | 0.012946 | standard |
| 3.49651 | 0.172747 | standard |
| 3.50762 | 0.143216 | standard |
| 3.97171 | 0.312784 | standard |
| 0.882652 | 1.0 | standard |
| 0.919549 | 0.9982 | standard |
| 2.38796 | 0.00491554 | standard |
| 2.39972 | 0.135178 | standard |
| 2.40578 | 0.314463 | standard |
| 2.41186 | 0.140043 | standard |
| 2.42356 | 0.00490041 | standard |
| 3.37967 | 0.144687 | standard |
| 3.38988 | 0.171683 | standard |
| 3.41749 | 0.0130942 | standard |
| 3.42277 | 0.163663 | standard |
| 3.42894 | 0.261385 | standard |
| 3.4351 | 0.16238 | standard |
| 3.44038 | 0.0129873 | standard |
| 3.49691 | 0.171411 | standard |
| 3.50712 | 0.144537 | standard |
| 3.97171 | 0.312889 | standard |
| 0.882652 | 1.0 | standard |
| 0.919549 | 0.999585 | standard |
| 2.39073 | 0.00493318 | standard |
| 2.40059 | 0.13851 | standard |
| 2.40581 | 0.315015 | standard |
| 2.41099 | 0.144169 | standard |
| 2.42089 | 0.00489506 | standard |
| 3.38056 | 0.146835 | standard |
| 3.38909 | 0.169675 | standard |
| 3.41924 | 0.012868 | standard |
| 3.42376 | 0.163746 | standard |
| 3.42891 | 0.263628 | standard |
| 3.43411 | 0.161895 | standard |
| 3.43862 | 0.0128957 | standard |
| 3.4977 | 0.169492 | standard |
| 3.50623 | 0.146723 | standard |
| 3.97171 | 0.313157 | standard |