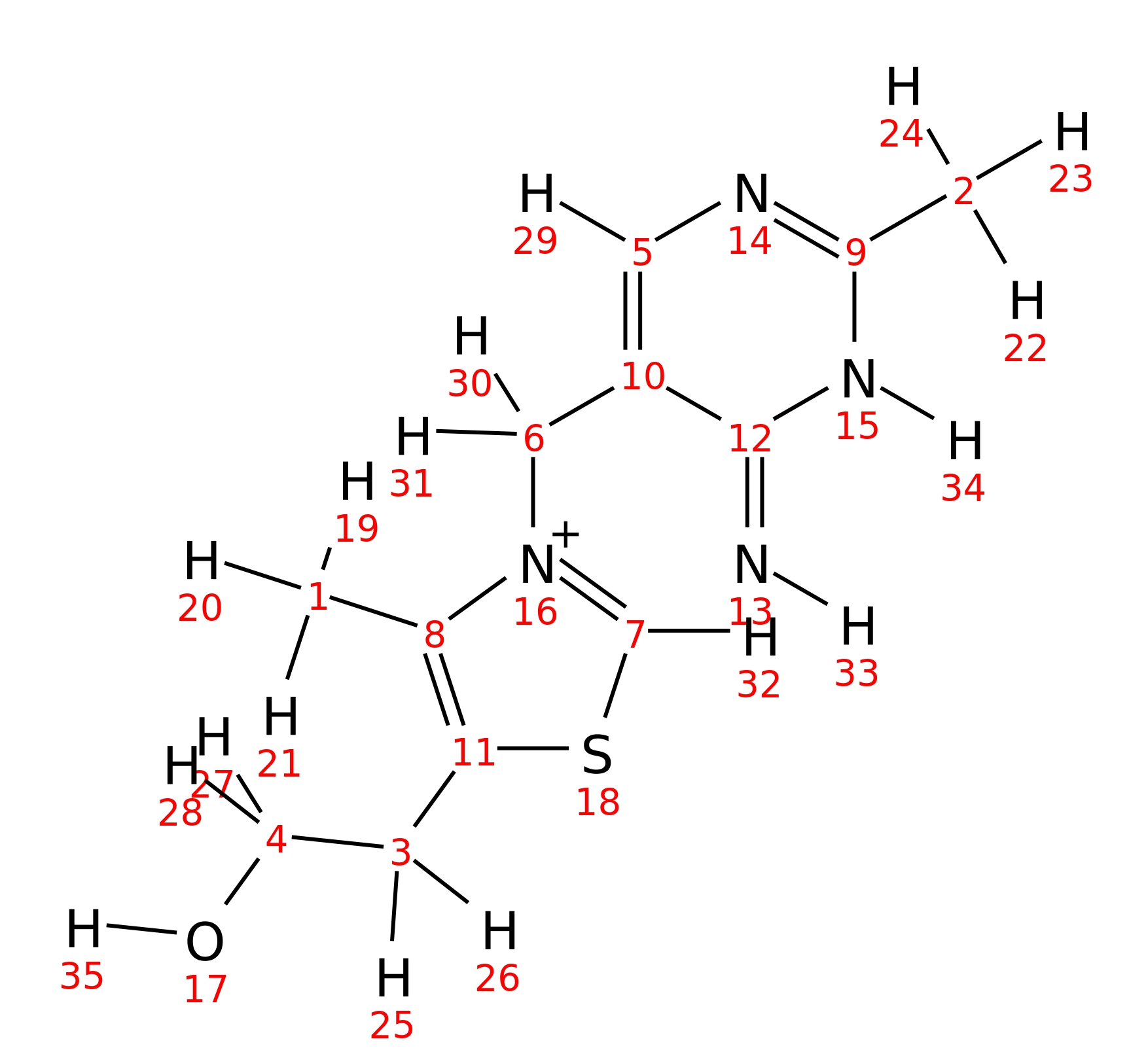
Thiamin
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.07637 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C12H17N4OS/c1-8-11(3-4-17)18-7-16(8)6-10-5-14-9(2)15-12(10)13/h5,7,17H,3-4,6H2,1-2H3,(H2,13,14,15)/q+1 | |
| Note 1 | Done but missing peak 32? | |
| Note 2 | 22,23,24?19,20,21 | |
| Note 3 | 25,26?27,28 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Thiamin | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 19 | 20 | 21 | 32 | 30 | 31 | 29 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 19 | 2.578 | -14.9 | -14.9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 2.578 | -14.9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 2.578 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 32 | 0 | 0 | 0 | 6.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 30 | 0 | 0 | 0 | 0 | 5.458 | -12.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 31 | 0 | 0 | 0 | 0 | 0 | 5.458 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 29 | 0 | 0 | 0 | 0 | 0 | 0 | 8.066 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.49 | -14.9 | -14.9 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.49 | -14.9 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.49 | 0 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.194 | -12.4 | 5.682 | 5.682 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.194 | 5.682 | 5.682 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.89 | -12.4 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.89 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.49007 | 0.999891 | standard |
| 2.57814 | 1.0 | standard |
| 3.17963 | 0.141676 | standard |
| 3.19374 | 0.272784 | standard |
| 3.20786 | 0.14966 | standard |
| 3.87611 | 0.14964 | standard |
| 3.89023 | 0.272762 | standard |
| 3.90434 | 0.141654 | standard |
| 5.45851 | 0.666203 | standard |
| 6.00007 | 0.331244 | standard |
| 8.06602 | 0.331529 | standard |
| 2.49056 | 0.999392 | standard |
| 2.57765 | 1.0 | standard |
| 3.04933 | 0.0844971 | standard |
| 3.18655 | 0.210721 | standard |
| 3.31832 | 0.176919 | standard |
| 3.76565 | 0.175998 | standard |
| 3.8974 | 0.208531 | standard |
| 4.03435 | 0.0807261 | standard |
| 5.45851 | 0.619793 | standard |
| 6.00007 | 0.309948 | standard |
| 8.06602 | 0.308452 | standard |
| 2.49017 | 0.999589 | standard |
| 2.57804 | 1.0 | standard |
| 3.09518 | 0.103003 | standard |
| 3.18849 | 0.239374 | standard |
| 3.28087 | 0.169453 | standard |
| 3.8031 | 0.168846 | standard |
| 3.89547 | 0.238262 | standard |
| 3.98879 | 0.101463 | standard |
| 5.45851 | 0.644002 | standard |
| 6.00007 | 0.320798 | standard |
| 8.06602 | 0.320484 | standard |
| 2.4901 | 0.999772 | standard |
| 2.57811 | 1.00001 | standard |
| 3.12033 | 0.114069 | standard |
| 3.19054 | 0.253884 | standard |
| 3.26063 | 0.165084 | standard |
| 3.82344 | 0.164731 | standard |
| 3.89346 | 0.253262 | standard |
| 3.96369 | 0.1132 | standard |
| 5.45851 | 0.653651 | standard |
| 6.00007 | 0.325514 | standard |
| 8.06602 | 0.325045 | standard |
| 2.49009 | 0.999904 | standard |
| 2.57812 | 1.0 | standard |
| 3.12873 | 0.117838 | standard |
| 3.19118 | 0.258104 | standard |
| 3.25362 | 0.163365 | standard |
| 3.83041 | 0.163248 | standard |
| 3.89281 | 0.257478 | standard |
| 3.95528 | 0.117131 | standard |
| 5.45851 | 0.656038 | standard |
| 6.00007 | 0.326446 | standard |
| 8.06602 | 0.326269 | standard |
| 2.49008 | 0.999575 | standard |
| 2.57813 | 1.0 | standard |
| 3.13543 | 0.12069 | standard |
| 3.19166 | 0.26104 | standard |
| 3.24799 | 0.162112 | standard |
| 3.83608 | 0.161893 | standard |
| 3.8923 | 0.260709 | standard |
| 3.94864 | 0.120213 | standard |
| 5.45851 | 0.658013 | standard |
| 6.00007 | 0.327608 | standard |
| 8.06602 | 0.327154 | standard |
| 2.49007 | 0.999872 | standard |
| 2.57814 | 1.0 | standard |
| 3.16509 | 0.133768 | standard |
| 3.19335 | 0.271577 | standard |
| 3.22158 | 0.154848 | standard |
| 3.86244 | 0.154583 | standard |
| 3.89067 | 0.271152 | standard |
| 3.91888 | 0.13372 | standard |
| 5.45851 | 0.664424 | standard |
| 6.00007 | 0.330815 | standard |
| 8.06602 | 0.330473 | standard |
| 2.49007 | 1.0 | standard |
| 2.57814 | 0.999862 | standard |
| 3.1748 | 0.137904 | standard |
| 3.19365 | 0.273407 | standard |
| 3.2125 | 0.151918 | standard |
| 3.87157 | 0.151988 | standard |
| 3.89035 | 0.273229 | standard |
| 3.90917 | 0.137905 | standard |
| 5.45851 | 0.665703 | standard |
| 6.00007 | 0.331438 | standard |
| 8.06602 | 0.331595 | standard |
| 2.49007 | 0.9997 | standard |
| 2.57814 | 1.0 | standard |
| 3.17963 | 0.141665 | standard |
| 3.19374 | 0.272722 | standard |
| 3.20791 | 0.149733 | standard |
| 3.87611 | 0.149631 | standard |
| 3.89023 | 0.272703 | standard |
| 3.90439 | 0.141712 | standard |
| 5.45851 | 0.665807 | standard |
| 6.00007 | 0.331435 | standard |
| 8.06602 | 0.331621 | standard |
| 2.49007 | 0.999564 | standard |
| 2.57814 | 1.0 | standard |
| 3.18254 | 0.143111 | standard |
| 3.19379 | 0.274328 | standard |
| 3.20514 | 0.147117 | standard |
| 3.87893 | 0.147315 | standard |
| 3.89018 | 0.274318 | standard |
| 3.90153 | 0.142886 | standard |
| 5.45851 | 0.666209 | standard |
| 6.00007 | 0.331656 | standard |
| 8.06602 | 0.331532 | standard |
| 2.49007 | 0.999759 | standard |
| 2.57814 | 1.0 | standard |
| 3.18442 | 0.144993 | standard |
| 3.19384 | 0.274569 | standard |
| 3.20327 | 0.144992 | standard |
| 3.88075 | 0.1452 | standard |
| 3.89018 | 0.274126 | standard |
| 3.89961 | 0.1452 | standard |
| 5.45851 | 0.666038 | standard |
| 6.00007 | 0.331407 | standard |
| 8.06602 | 0.331547 | standard |
| 2.49007 | 0.999876 | standard |
| 2.57814 | 1.0 | standard |
| 3.18575 | 0.145118 | standard |
| 3.19384 | 0.274452 | standard |
| 3.20194 | 0.145118 | standard |
| 3.88209 | 0.145367 | standard |
| 3.89013 | 0.274517 | standard |
| 3.89822 | 0.145121 | standard |
| 5.45851 | 0.665906 | standard |
| 6.00007 | 0.331299 | standard |
| 8.06602 | 0.33123 | standard |
| 2.49007 | 0.99987 | standard |
| 2.57814 | 1.0 | standard |
| 3.18634 | 0.145322 | standard |
| 3.19384 | 0.27453 | standard |
| 3.20144 | 0.145008 | standard |
| 3.88263 | 0.145316 | standard |
| 3.89013 | 0.274524 | standard |
| 3.89773 | 0.145002 | standard |
| 5.45851 | 0.66644 | standard |
| 6.00007 | 0.33148 | standard |
| 8.06602 | 0.331615 | standard |
| 2.49007 | 0.999883 | standard |
| 2.57814 | 1.0 | standard |
| 3.18684 | 0.145314 | standard |
| 3.19387 | 0.27464 | standard |
| 3.20095 | 0.145148 | standard |
| 3.88307 | 0.145343 | standard |
| 3.89013 | 0.274482 | standard |
| 3.89724 | 0.145047 | standard |
| 5.45851 | 0.665967 | standard |
| 6.00007 | 0.331649 | standard |
| 8.06602 | 0.331865 | standard |
| 2.49007 | 0.999881 | standard |
| 2.57814 | 1.0 | standard |
| 3.18762 | 0.145224 | standard |
| 3.19389 | 0.274873 | standard |
| 3.20016 | 0.145222 | standard |
| 3.88386 | 0.145371 | standard |
| 3.89013 | 0.274447 | standard |
| 3.89645 | 0.14502 | standard |
| 5.45851 | 0.666266 | standard |
| 6.00007 | 0.331863 | standard |
| 8.06602 | 0.331479 | standard |
| 2.49007 | 1.0 | standard |
| 2.57814 | 0.999763 | standard |
| 3.18792 | 0.145025 | standard |
| 3.19389 | 0.274693 | standard |
| 3.19986 | 0.145025 | standard |
| 3.88421 | 0.145237 | standard |
| 3.89013 | 0.274421 | standard |
| 3.8961 | 0.145219 | standard |
| 5.45851 | 0.666384 | standard |
| 6.00007 | 0.331313 | standard |
| 8.06602 | 0.331451 | standard |
| 2.49007 | 0.999868 | standard |
| 2.57814 | 1.0 | standard |
| 3.18822 | 0.145023 | standard |
| 3.19389 | 0.274734 | standard |
| 3.19957 | 0.145023 | standard |
| 3.88451 | 0.145247 | standard |
| 3.89013 | 0.274428 | standard |
| 3.89581 | 0.145237 | standard |
| 5.45851 | 0.666719 | standard |
| 6.00007 | 0.331514 | standard |
| 8.06602 | 0.331437 | standard |
| 2.49007 | 0.999632 | standard |
| 2.57814 | 1.0 | standard |
| 3.18876 | 0.145506 | standard |
| 3.19389 | 0.274538 | standard |
| 3.19902 | 0.145506 | standard |
| 3.885 | 0.145261 | standard |
| 3.89013 | 0.274047 | standard |
| 3.89526 | 0.145262 | standard |
| 5.45851 | 0.666436 | standard |
| 6.00007 | 0.331425 | standard |
| 8.06602 | 0.331854 | standard |
| 2.49007 | 0.999533 | standard |
| 2.57814 | 1.0 | standard |
| 3.18955 | 0.145688 | standard |
| 3.19389 | 0.27428 | standard |
| 3.19823 | 0.145688 | standard |
| 3.88579 | 0.145345 | standard |
| 3.89013 | 0.273594 | standard |
| 3.89447 | 0.145348 | standard |
| 5.45851 | 0.666523 | standard |
| 6.00007 | 0.331975 | standard |
| 8.06602 | 0.331964 | standard |