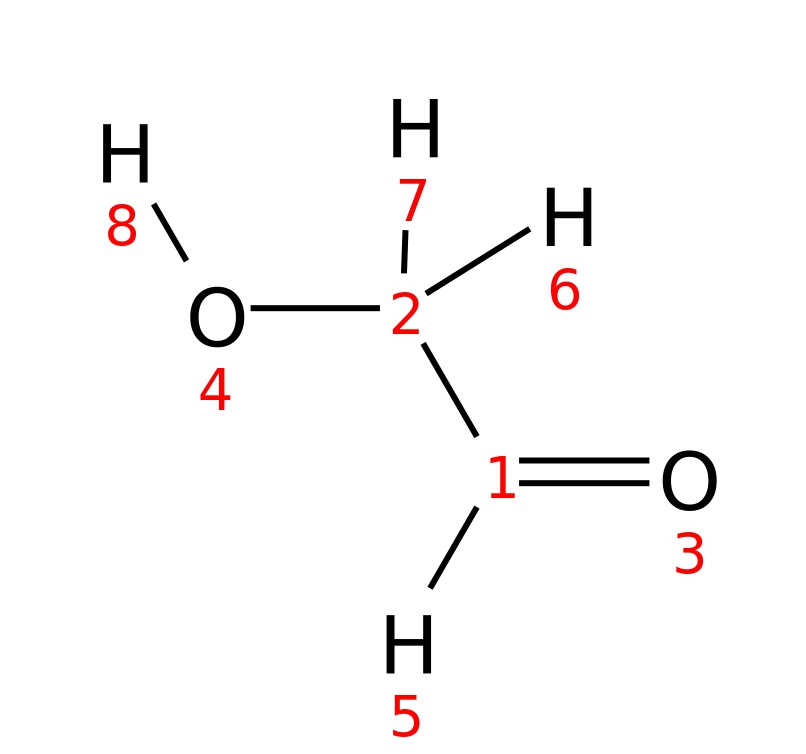
Glycolaldehyde
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.00886 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C2H4O2/c3-1-2-4/h1,4H,2H2 | |
| Note 1 | hydrate |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Glycolaldehyde | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 6 | 7 | 5 | |
|---|---|---|---|
| 6 | 3.505 | -12.4 | 3.466 |
| 7 | 0 | 3.505 | 6.846 |
| 5 | 0 | 0 | 5.045 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.49851 | 0.995383 | standard |
| 3.51137 | 1.00004 | standard |
| 5.03274 | 0.262426 | standard |
| 5.04555 | 0.493995 | standard |
| 5.05846 | 0.262168 | standard |
| 3.43797 | 0.83073 | standard |
| 3.56614 | 1.0 | standard |
| 4.74244 | 0.0154701 | standard |
| 4.92321 | 0.282942 | standard |
| 5.0514 | 0.435528 | standard |
| 5.17967 | 0.208549 | standard |
| 3.4609 | 0.885657 | standard |
| 3.5465 | 1.0 | standard |
| 4.84272 | 0.0142091 | standard |
| 4.96249 | 0.275562 | standard |
| 5.04816 | 0.458293 | standard |
| 5.13371 | 0.224697 | standard |
| 3.47211 | 0.913929 | standard |
| 3.53635 | 1.00001 | standard |
| 4.98278 | 0.27207 | standard |
| 5.04698 | 0.468982 | standard |
| 5.11127 | 0.233395 | standard |
| 3.47587 | 0.922691 | standard |
| 3.53299 | 1.0 | standard |
| 4.98961 | 0.270907 | standard |
| 5.04673 | 0.472056 | standard |
| 5.10375 | 0.236476 | standard |
| 3.47878 | 0.929718 | standard |
| 3.53022 | 1.0 | standard |
| 4.99505 | 0.270026 | standard |
| 5.04644 | 0.474596 | standard |
| 5.09782 | 0.238877 | standard |
| 3.49198 | 0.970177 | standard |
| 3.5177 | 1.00004 | standard |
| 5.02008 | 0.264415 | standard |
| 5.04575 | 0.486593 | standard |
| 5.07151 | 0.253735 | standard |
| 3.49638 | 0.996496 | standard |
| 3.5135 | 1.00006 | standard |
| 5.02849 | 0.262442 | standard |
| 5.0456 | 0.49387 | standard |
| 5.06281 | 0.262248 | standard |
| 3.49851 | 0.995386 | standard |
| 3.51137 | 1.00005 | standard |
| 5.03274 | 0.262432 | standard |
| 5.04555 | 0.493994 | standard |
| 5.05846 | 0.262174 | standard |
| 3.49985 | 0.996993 | standard |
| 3.51008 | 1.00005 | standard |
| 5.03531 | 0.262605 | standard |
| 5.04555 | 0.494151 | standard |
| 5.05589 | 0.262281 | standard |
| 3.50069 | 0.99786 | standard |
| 3.50924 | 1.00049 | standard |
| 5.037 | 0.262291 | standard |
| 5.04555 | 0.493663 | standard |
| 5.05411 | 0.262291 | standard |
| 3.50128 | 0.997191 | standard |
| 3.50865 | 1.00073 | standard |
| 5.03818 | 0.262339 | standard |
| 5.04555 | 0.493779 | standard |
| 5.05292 | 0.262339 | standard |
| 3.50153 | 1.00086 | standard |
| 3.5084 | 0.996941 | standard |
| 5.03868 | 0.262445 | standard |
| 5.04555 | 0.493873 | standard |
| 5.05243 | 0.262445 | standard |
| 3.50177 | 0.996412 | standard |
| 3.50815 | 1.00099 | standard |
| 5.03907 | 0.26242 | standard |
| 5.0455 | 0.494407 | standard |
| 5.05193 | 0.26242 | standard |
| 3.50212 | 1.00128 | standard |
| 3.50786 | 1.00133 | standard |
| 5.03987 | 0.263311 | standard |
| 5.0455 | 0.495343 | standard |
| 5.05124 | 0.26272 | standard |
| 3.50227 | 0.995081 | standard |
| 3.50766 | 1.00149 | standard |
| 5.04016 | 0.263258 | standard |
| 5.0455 | 0.495231 | standard |
| 5.05094 | 0.262636 | standard |
| 3.50242 | 1.00163 | standard |
| 3.50756 | 1.00168 | standard |
| 5.04036 | 0.262974 | standard |
| 5.0455 | 0.495451 | standard |
| 5.05065 | 0.262974 | standard |
| 3.50262 | 1.00202 | standard |
| 3.50726 | 1.00206 | standard |
| 5.04085 | 0.263281 | standard |
| 5.0455 | 0.495545 | standard |
| 5.05025 | 0.262579 | standard |
| 3.50301 | 1.00293 | standard |
| 3.50697 | 1.00297 | standard |
| 5.04155 | 0.263764 | standard |
| 5.0455 | 0.496941 | standard |
| 5.04946 | 0.263764 | standard |