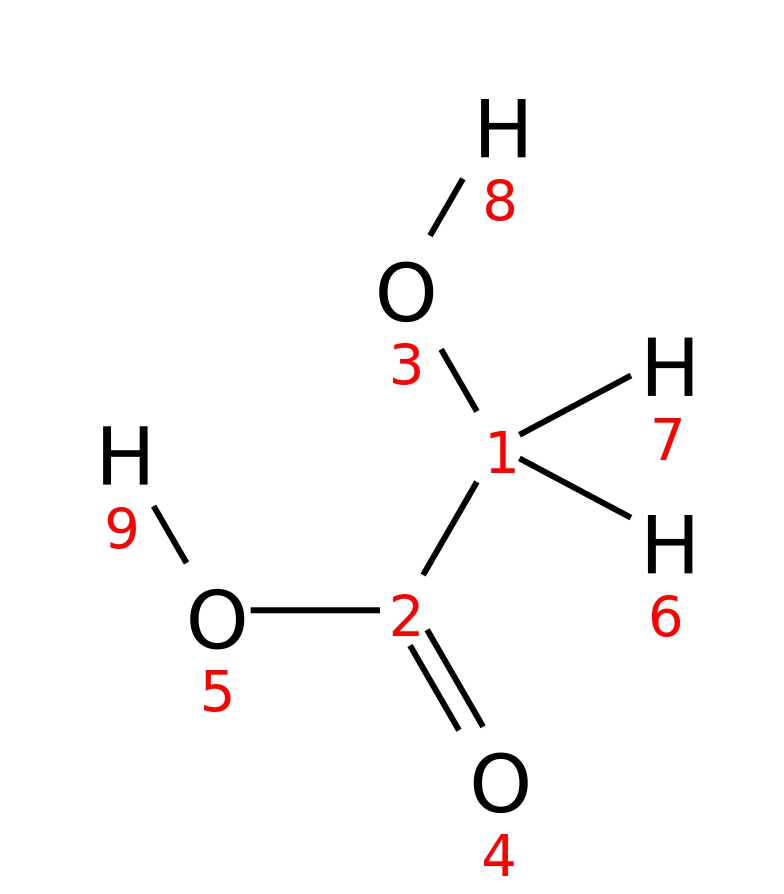
Glycolate
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.00978 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C2H4O3/c3-1-2(4)5/h3H,1H2,(H,4,5) | |
| pKa | 3.83 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Glycolate | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 6 | 7 | |
|---|---|---|
| 6 | 3.928 | -12.4 |
| 7 | 0 | 3.928 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |
| 3.92797 | 1.0 | standard |