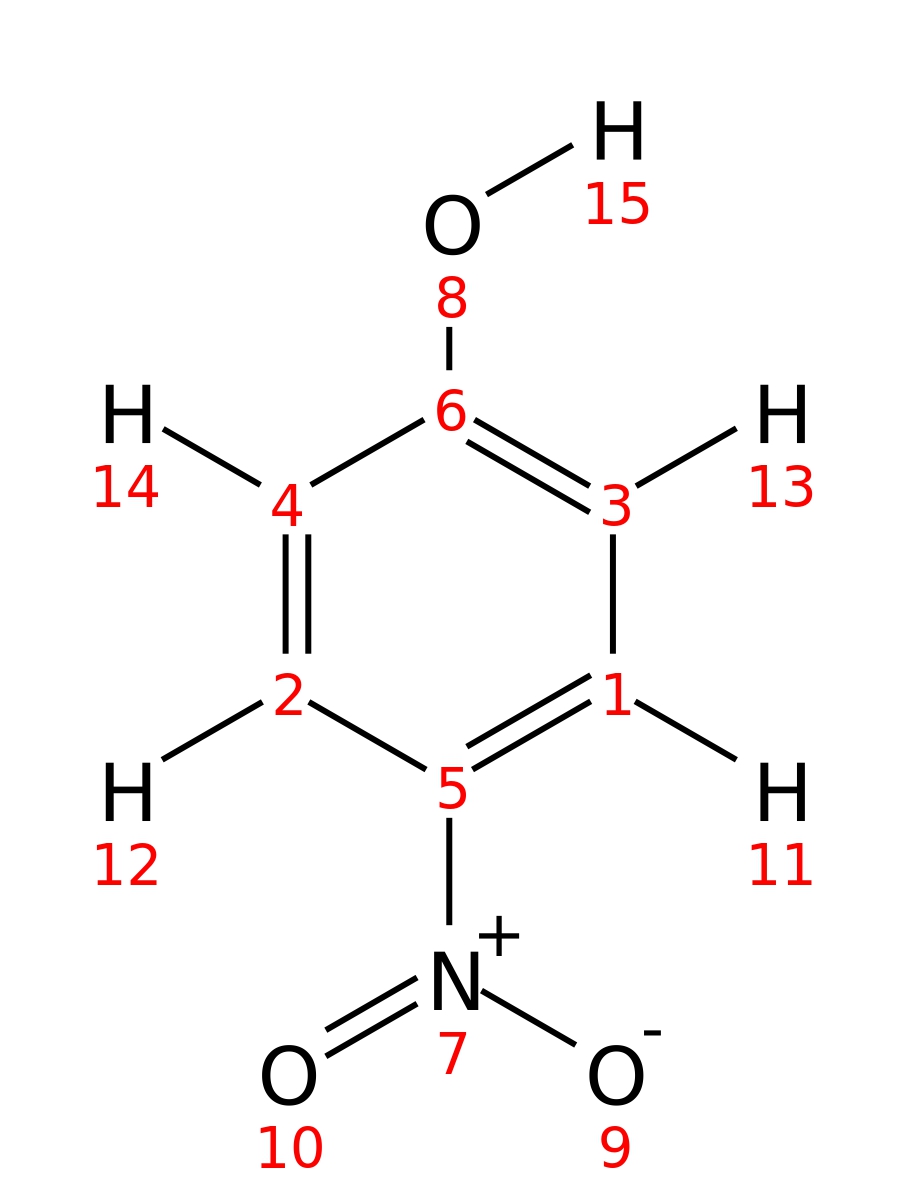
4-Nitrophenol
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.02111 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H5NO3/c8-6-3-1-5(2-4-6)7(9)10/h1-4,8H | |
| Note 1 | 11 and 12 non equivalent (strange) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4-Nitrophenol | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 11 | 13 | 14 | 12 | |
|---|---|---|---|---|
| 11 | 8.045 | 8.832 | 0 | 2.143 |
| 13 | 0 | 6.678 | 2.143 | 0 |
| 14 | 0 | 0 | 6.678 | 8.832 |
| 12 | 0 | 0 | 0 | 8.056 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| -0.987755 | 1.125e-06 | standard |
| 6.66706 | 0.997912 | standard |
| 6.68855 | 1.00005 | standard |
| 8.0365 | 0.467817 | standard |
| 8.04339 | 0.541805 | standard |
| 8.04998 | 0.423708 | standard |
| 8.05148 | 0.424972 | standard |
| 8.05784 | 0.54544 | standard |
| 8.06486 | 0.466997 | standard |
| -0.986465 | 4.1125e-05 | standard |
| 6.5595 | 0.736863 | standard |
| 6.77881 | 1.0 | standard |
| 7.94965 | 0.99204 | standard |
| 8.16899 | 0.730943 | standard |
| -0.963454 | 1.9125e-05 | standard |
| 6.6012 | 0.815244 | standard |
| 6.74678 | 1.0 | standard |
| 7.98174 | 0.984499 | standard |
| 8.12721 | 0.802852 | standard |
| -0.944509 | 1.1125e-05 | standard |
| 6.62131 | 0.858104 | standard |
| 6.73011 | 1.0 | standard |
| 7.99844 | 0.971398 | standard |
| 8.10698 | 0.833757 | standard |
| -0.95423 | 9.125e-06 | standard |
| 6.62791 | 0.87015 | standard |
| 6.72447 | 1.00001 | standard |
| 8.00423 | 0.963097 | standard |
| 8.10036 | 0.838022 | standard |
| -0.900272 | 7.125e-06 | standard |
| 6.63308 | 0.882798 | standard |
| 6.71988 | 1.0 | standard |
| 8.00885 | 0.953721 | standard |
| 8.09507 | 0.842213 | standard |
| -0.791663 | 2.125e-06 | standard |
| 6.65597 | 0.941315 | standard |
| 6.69917 | 1.00003 | standard |
| 8.03132 | 0.818923 | standard |
| 8.07047 | 0.774699 | standard |
| -0.575933 | 1.125e-06 | standard |
| 6.66336 | 0.97577 | standard |
| 6.6779 | 0.189173 | standard |
| 6.69215 | 1.00012 | standard |
| 8.03389 | 0.561706 | standard |
| 8.03873 | 0.656581 | standard |
| 8.06266 | 0.644891 | standard |
| 8.06743 | 0.547876 | standard |
| -0.987259 | 1.125e-06 | standard |
| 6.66708 | 1.00005 | standard |
| 6.68855 | 1.00005 | standard |
| 8.0365 | 0.467917 | standard |
| 8.04339 | 0.541898 | standard |
| 8.04998 | 0.423779 | standard |
| 8.05148 | 0.425041 | standard |
| 8.05784 | 0.545531 | standard |
| 8.06486 | 0.467097 | standard |
| 6.6692 | 0.993353 | standard |
| 6.67788 | 0.209592 | standard |
| 6.68642 | 1.00005 | standard |
| 8.03797 | 0.42423 | standard |
| 8.04597 | 0.518276 | standard |
| 8.0506 | 0.579947 | standard |
| 8.05517 | 0.523418 | standard |
| 8.06323 | 0.423829 | standard |
| 6.67064 | 1.00015 | standard |
| 6.68508 | 1.00015 | standard |
| 8.03927 | 0.403178 | standard |
| 8.04779 | 0.5617 | standard |
| 8.05055 | 0.645781 | standard |
| 8.05331 | 0.561541 | standard |
| 8.06199 | 0.403772 | standard |
| 6.6717 | 0.993131 | standard |
| 6.68401 | 1.00074 | standard |
| 8.04 | 0.38346 | standard |
| 8.04912 | 0.699491 | standard |
| 8.05208 | 0.699649 | standard |
| 8.06109 | 0.382708 | standard |
| 6.67206 | 1.0 | standard |
| 6.68366 | 0.999196 | standard |
| 8.04043 | 0.380864 | standard |
| 8.04923 | 0.757413 | standard |
| 8.05197 | 0.757545 | standard |
| 8.06064 | 0.376224 | standard |
| 6.6724 | 0.997387 | standard |
| 6.68333 | 1.00063 | standard |
| 8.04075 | 0.372404 | standard |
| 8.04931 | 0.776354 | standard |
| 8.05182 | 0.776577 | standard |
| 8.06035 | 0.372524 | standard |
| 6.67295 | 1.0 | standard |
| 6.68277 | 0.99677 | standard |
| 8.04122 | 0.379736 | standard |
| 8.04968 | 0.748145 | standard |
| 8.05142 | 0.748145 | standard |
| 8.05988 | 0.379736 | standard |
| 6.67327 | 1.00031 | standard |
| 6.68256 | 0.98526 | standard |
| 8.04142 | 0.38267 | standard |
| 8.05055 | 0.73392 | standard |
| 8.05968 | 0.382698 | standard |
| 6.67348 | 1.0006 | standard |
| 6.68222 | 0.995746 | standard |
| 8.04163 | 0.378264 | standard |
| 8.05055 | 0.749355 | standard |
| 8.05947 | 0.377977 | standard |
| 6.6739 | 1.00102 | standard |
| 6.68181 | 0.988993 | standard |
| 8.04109 | 0.357469 | standard |
| 8.04193 | 0.369759 | standard |
| 8.05055 | 0.627223 | standard |
| 8.05917 | 0.369759 | standard |
| 8.06001 | 0.357469 | standard |
| 6.67463 | 0.993916 | standard |
| 6.6812 | 1.00023 | standard |
| 8.04243 | 0.3645 | standard |
| 8.04903 | 0.40525 | standard |
| 8.05207 | 0.405297 | standard |
| 8.0587 | 0.357947 | standard |
| 8.0593 | 0.350288 | standard |