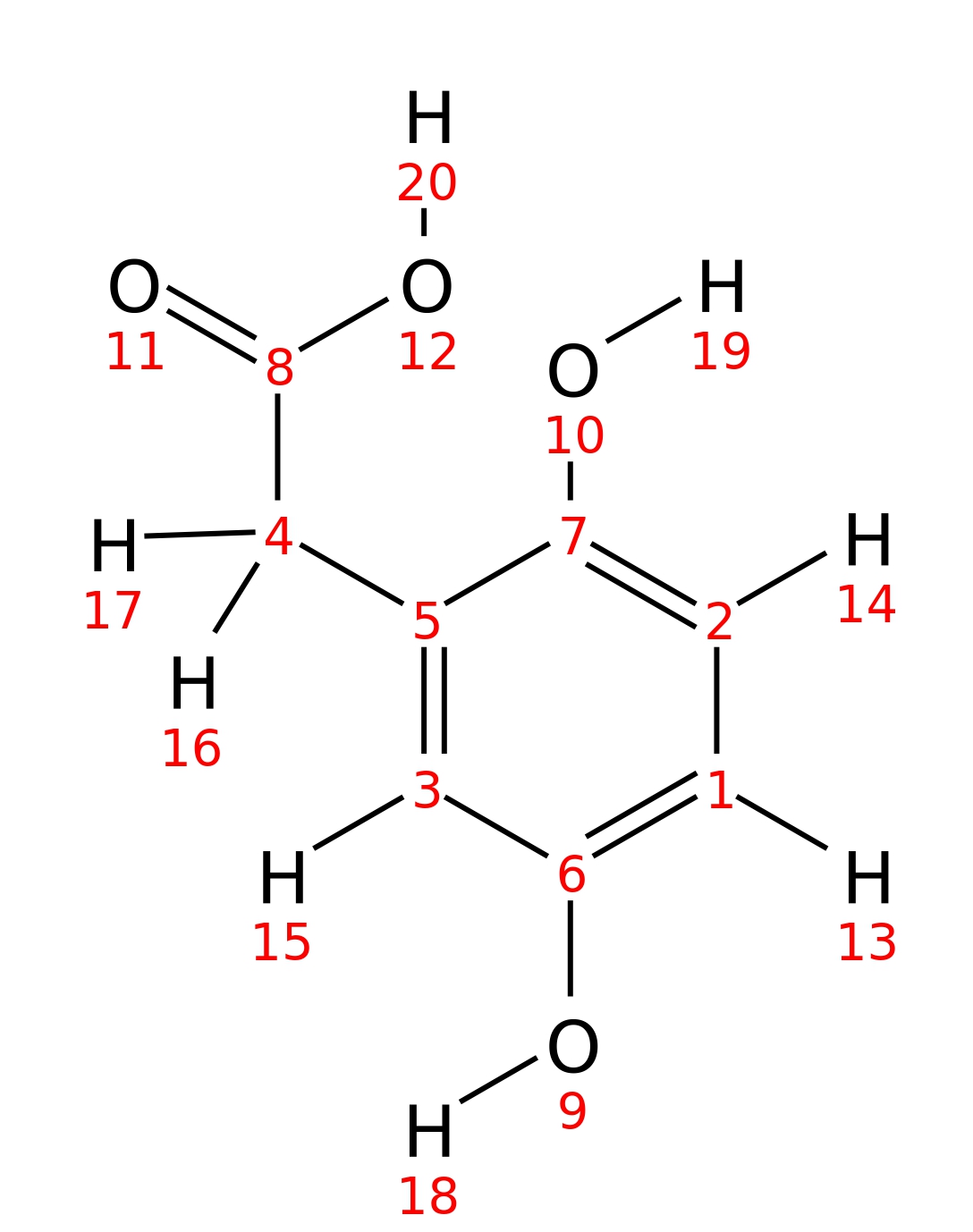
Homogentisic-acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.01043 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H8O4/c9-6-1-2-7(10)5(3-6)4-8(11)12/h1-3,9-10H,4H2,(H,11,12) | |
| Note 1 | broad |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Homogentisate | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|
| 13 | 6.694 | 8.62 | 2.201 | 0 | 0 |
| 14 | 0 | 6.797 | 0 | 0 | 0 |
| 15 | 0 | 0 | 6.706 | 0 | 0 |
| 16 | 0 | 0 | 0 | 3.471 | -12.4 |
| 17 | 0 | 0 | 0 | 0 | 3.471 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.47141 | 1.0 | standard |
| 6.67916 | 0.105824 | standard |
| 6.68418 | 0.149427 | standard |
| 6.70435 | 0.674633 | standard |
| 6.78778 | 0.255049 | standard |
| 6.80819 | 0.172161 | standard |
| 3.47141 | 0.948025 | standard |
| 6.53969 | 0.0405321 | standard |
| 6.72609 | 1.0 | standard |
| 6.97949 | 0.0322611 | standard |
| 3.47141 | 1.0 | standard |
| 6.59818 | 0.059154 | standard |
| 6.71947 | 0.84662 | standard |
| 6.75501 | 0.615182 | standard |
| 6.87683 | 0.0380061 | standard |
| 6.91055 | 0.0503933 | standard |
| 3.47141 | 1.0 | standard |
| 6.62684 | 0.075512 | standard |
| 6.71647 | 0.761315 | standard |
| 6.762 | 0.462088 | standard |
| 6.85213 | 0.0427941 | standard |
| 6.87732 | 0.0668277 | standard |
| 3.47141 | 1.0 | standard |
| 6.63604 | 0.0829851 | standard |
| 6.71536 | 0.73685 | standard |
| 6.76446 | 0.422086 | standard |
| 6.84454 | 0.045167 | standard |
| 6.86665 | 0.0742411 | standard |
| 3.47141 | 1.0 | standard |
| 6.6432 | 0.089856 | standard |
| 6.71442 | 0.719828 | standard |
| 6.76655 | 0.393782 | standard |
| 6.83894 | 0.0475951 | standard |
| 6.85833 | 0.081276 | standard |
| 3.47141 | 1.0 | standard |
| 6.66294 | 0.0840501 | standard |
| 6.67229 | 0.129931 | standard |
| 6.70856 | 0.680483 | standard |
| 6.77856 | 0.27086 | standard |
| 6.82365 | 0.119583 | standard |
| 3.47141 | 1.0 | standard |
| 6.67391 | 0.0977284 | standard |
| 6.68041 | 0.143834 | standard |
| 6.70583 | 0.683815 | standard |
| 6.78461 | 0.234541 | standard |
| 6.81295 | 0.139099 | standard |
| 3.47141 | 1.0 | standard |
| 6.67916 | 0.105832 | standard |
| 6.68418 | 0.149434 | standard |
| 6.70435 | 0.674731 | standard |
| 6.78778 | 0.254838 | standard |
| 6.80819 | 0.172146 | standard |
| 3.47141 | 1.0 | standard |
| 6.68224 | 0.11188 | standard |
| 6.68627 | 0.151989 | standard |
| 6.70371 | 0.583033 | standard |
| 6.78933 | 0.273959 | standard |
| 6.80588 | 0.197812 | standard |
| 3.47141 | 1.0 | standard |
| 6.68426 | 0.115993 | standard |
| 6.68767 | 0.153101 | standard |
| 6.70416 | 0.443269 | standard |
| 6.79039 | 0.279152 | standard |
| 6.80437 | 0.21277 | standard |
| 3.47141 | 1.0 | standard |
| 6.68572 | 0.120481 | standard |
| 6.68856 | 0.154421 | standard |
| 6.69805 | 0.138521 | standard |
| 6.70081 | 0.266554 | standard |
| 6.70442 | 0.391451 | standard |
| 6.70719 | 0.244209 | standard |
| 6.79123 | 0.277835 | standard |
| 6.80328 | 0.221121 | standard |
| 3.47141 | 1.0 | standard |
| 6.68619 | 0.120409 | standard |
| 6.68897 | 0.153703 | standard |
| 6.69766 | 0.13812 | standard |
| 6.70036 | 0.246122 | standard |
| 6.70449 | 0.374638 | standard |
| 6.70712 | 0.248117 | standard |
| 6.79158 | 0.278012 | standard |
| 6.80288 | 0.224736 | standard |
| 3.47141 | 1.0 | standard |
| 6.68667 | 0.121928 | standard |
| 6.68927 | 0.153966 | standard |
| 6.6974 | 0.138715 | standard |
| 6.69993 | 0.231544 | standard |
| 6.70454 | 0.36524 | standard |
| 6.70706 | 0.248529 | standard |
| 6.79188 | 0.276563 | standard |
| 6.80249 | 0.22642 | standard |
| 3.47141 | 1.0 | standard |
| 6.68756 | 0.125794 | standard |
| 6.68976 | 0.155121 | standard |
| 6.69702 | 0.139566 | standard |
| 6.69931 | 0.212959 | standard |
| 6.70469 | 0.354372 | standard |
| 6.70686 | 0.257811 | standard |
| 6.79237 | 0.273848 | standard |
| 6.80184 | 0.231228 | standard |
| 3.47141 | 1.0 | standard |
| 6.68783 | 0.124532 | standard |
| 6.69007 | 0.153729 | standard |
| 6.69692 | 0.141999 | standard |
| 6.69901 | 0.207543 | standard |
| 6.70469 | 0.348689 | standard |
| 6.70678 | 0.258408 | standard |
| 6.79262 | 0.274387 | standard |
| 6.80154 | 0.232338 | standard |
| 3.47141 | 1.0 | standard |
| 6.68812 | 0.125071 | standard |
| 6.69027 | 0.153439 | standard |
| 6.6968 | 0.142065 | standard |
| 6.69881 | 0.202124 | standard |
| 6.70479 | 0.344002 | standard |
| 6.70679 | 0.259337 | standard |
| 6.79282 | 0.273318 | standard |
| 6.80135 | 0.233246 | standard |
| 3.47141 | 1.0 | standard |
| 6.68872 | 0.128605 | standard |
| 6.69056 | 0.15477 | standard |
| 6.69647 | 0.143058 | standard |
| 6.69831 | 0.194213 | standard |
| 6.70483 | 0.33791 | standard |
| 6.70667 | 0.26246 | standard |
| 6.79312 | 0.271418 | standard |
| 6.8009 | 0.235526 | standard |
| 3.47141 | 1.0 | standard |
| 6.68949 | 0.131041 | standard |
| 6.69106 | 0.154517 | standard |
| 6.69603 | 0.141673 | standard |
| 6.69772 | 0.182254 | standard |
| 6.70493 | 0.328904 | standard |
| 6.70649 | 0.26684 | standard |
| 6.79366 | 0.269394 | standard |
| 6.80031 | 0.238124 | standard |