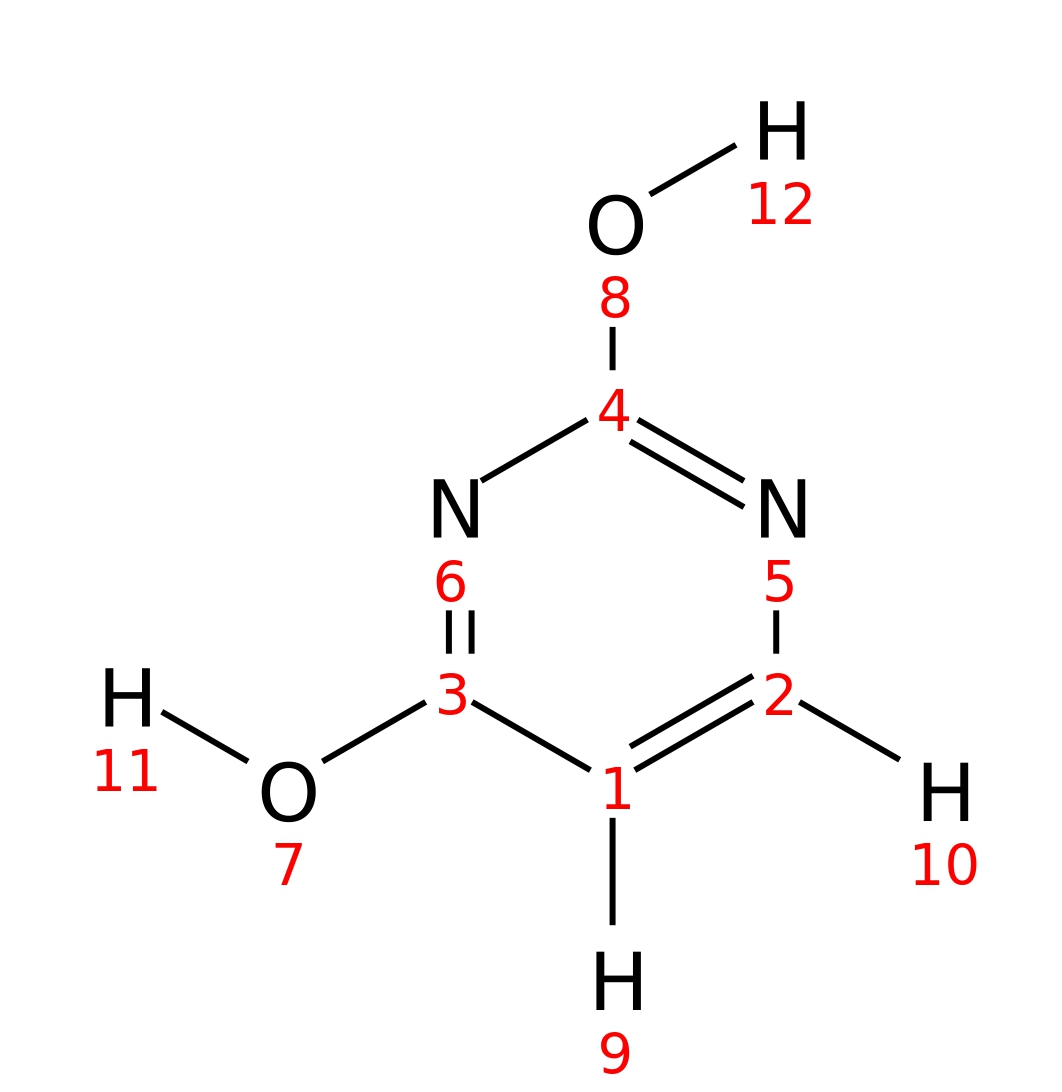
Uracil
Simulation outputs:
|
Parameter | Value |
| Field strength | 499.84(MHz) | |
| RMSD of the fit | 0.01065 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C4H4N2O2/c7-3-1-2-5-4(8)6-3/h1-2H,(H2,5,6,7,8) | |
| pKa | 9.45 | |
| Note 1 | 9,10 assigned using qm |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Uracil | Solute | saturated solutionmM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 1% |
Spin System Matrix
| 9 | 10 | |
|---|---|---|
| 9 | 5.792 | 7.6 |
| 10 | 0 | 7.527 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 5.78434 | 0.99994 | standard |
| 5.79962 | 0.99994 | standard |
| 7.51901 | 1.0 | standard |
| 7.53419 | 1.0 | standard |
| -0.829751 | 3.125e-06 | standard |
| 5.69181 | 0.805231 | standard |
| 5.88184 | 1.0 | standard |
| 7.43669 | 1.0 | standard |
| 7.62672 | 0.805231 | standard |
| -0.963553 | 2.125e-06 | standard |
| 5.72642 | 0.865386 | standard |
| 5.85307 | 1.0 | standard |
| 7.46555 | 1.0 | standard |
| 7.59221 | 0.865386 | standard |
| -0.708446 | 1.125e-06 | standard |
| 5.74318 | 0.897198 | standard |
| 5.8382 | 1.0 | standard |
| 7.48033 | 1.0 | standard |
| 7.57535 | 0.897198 | standard |
| -0.809814 | 1.125e-06 | standard |
| 5.74874 | 0.908068 | standard |
| 5.83324 | 1.0 | standard |
| 7.48529 | 1.0 | standard |
| 7.56979 | 0.908068 | standard |
| -0.909794 | 1.125e-06 | standard |
| 5.7532 | 0.916863 | standard |
| 5.82917 | 1.0 | standard |
| 7.48936 | 1.0 | standard |
| 7.56533 | 0.916863 | standard |
| 5.77284 | 0.957519 | standard |
| 5.81082 | 1.0 | standard |
| 7.5077 | 1.0 | standard |
| 7.54569 | 0.957519 | standard |
| 5.77928 | 1.0 | standard |
| 5.80457 | 1.0 | standard |
| 7.51395 | 1.0 | standard |
| 7.53924 | 1.0 | standard |
| 5.78246 | 0.999952 | standard |
| 5.8015 | 0.999952 | standard |
| 7.51713 | 1.0 | standard |
| 7.53607 | 1.0 | standard |
| 5.78444 | 1.0 | standard |
| 5.79962 | 1.0 | standard |
| 7.51901 | 1.0 | standard |
| 7.53419 | 1.0 | standard |
| 5.78563 | 1.0 | standard |
| 5.79833 | 1.0 | standard |
| 7.5202 | 1.0 | standard |
| 7.5329 | 1.0 | standard |
| 5.78662 | 1.0 | standard |
| 5.79743 | 1.0 | standard |
| 7.52109 | 0.999916 | standard |
| 7.532 | 0.999916 | standard |
| 5.78692 | 1.0 | standard |
| 5.79704 | 1.0 | standard |
| 7.52149 | 1.0 | standard |
| 7.53161 | 1.0 | standard |
| 5.78722 | 1.0 | standard |
| 5.79674 | 1.0 | standard |
| 7.52179 | 1.0 | standard |
| 7.53131 | 1.0 | standard |
| 5.78781 | 1.0 | standard |
| 5.79624 | 1.0 | standard |
| 7.52228 | 0.999894 | standard |
| 7.53081 | 0.999894 | standard |
| 5.78801 | 0.999886 | standard |
| 5.79604 | 0.999886 | standard |
| 7.52258 | 1.0 | standard |
| 7.53052 | 1.0 | standard |
| 5.78821 | 0.99988 | standard |
| 5.79585 | 0.99988 | standard |
| 7.52278 | 1.0 | standard |
| 7.53032 | 1.0 | standard |
| 5.78861 | 1.0 | standard |
| 5.79545 | 1.0 | standard |
| 7.52308 | 0.999868 | standard |
| 7.53002 | 0.999868 | standard |
| 5.7891 | 1.0 | standard |
| 5.79495 | 1.0 | standard |
| 7.52357 | 1.0 | standard |
| 7.52943 | 1.0 | standard |