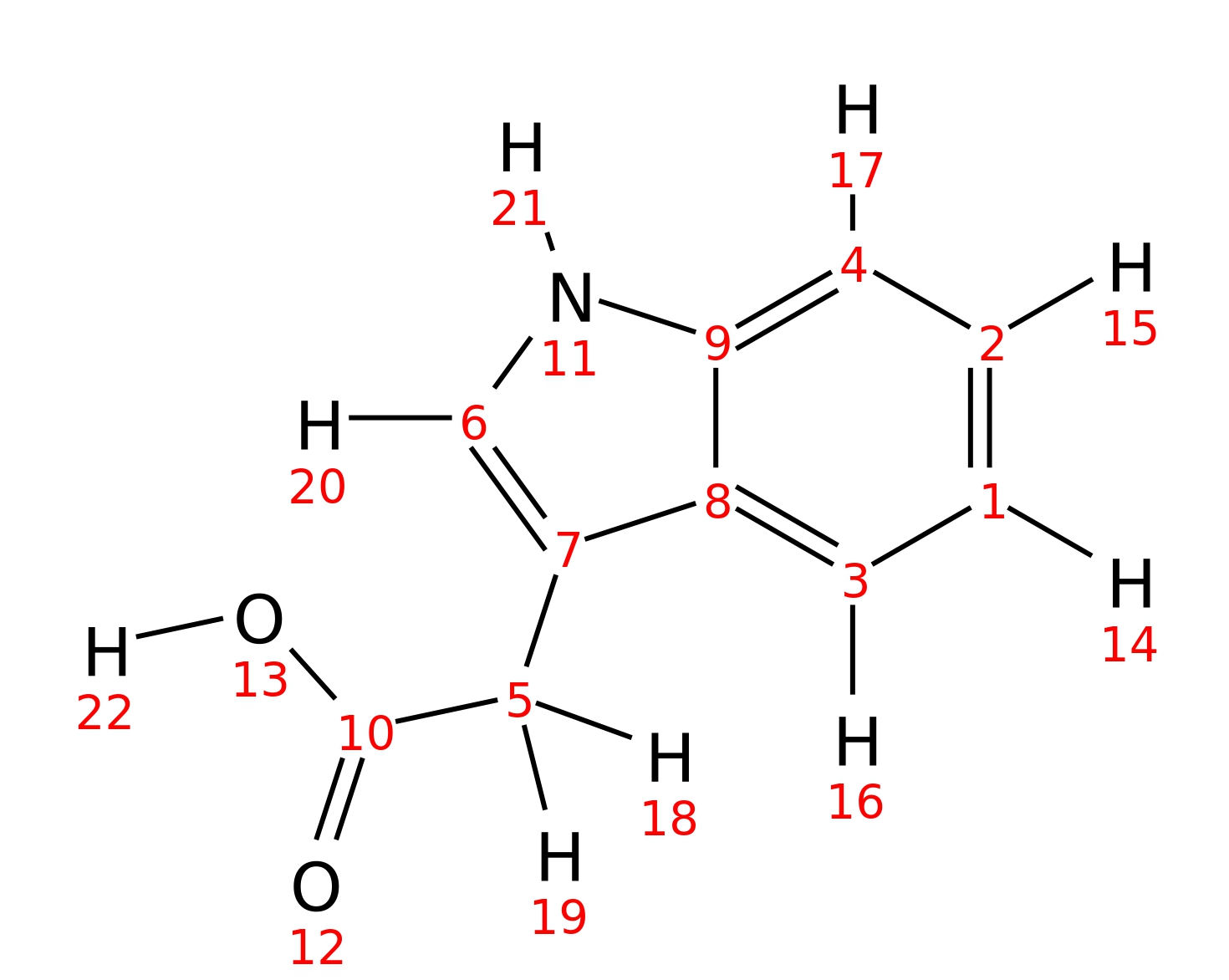
Indole-3-acetic-acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.01586 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H9NO2/c12-10(13)5-7-6-11-9-4-2-1-3-8(7)9/h1-4,6,11H,5H2,(H,12,13) | |
| Note 1 | 15,17?14,16 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Indole-3-acetate | Solute | Saturated1 |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 15 | 14 | 16 | 17 | 20 | 18 | 19 | |
|---|---|---|---|---|---|---|---|
| 15 | 7.155 | 7.5 | 0 | 7.5 | 0 | 0 | 0 |
| 14 | 0 | 7.238 | 7.5 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 7.63 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 7.49 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 7.223 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 3.652 | -12.4 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 3.652 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.65233 | 1.0 | standard |
| 7.13529 | 0.101614 | standard |
| 7.15391 | 0.248417 | standard |
| 7.17249 | 0.172893 | standard |
| 7.22278 | 0.595401 | standard |
| 7.23882 | 0.269397 | standard |
| 7.25741 | 0.116884 | standard |
| 7.48106 | 0.270356 | standard |
| 7.49953 | 0.24349 | standard |
| 7.62101 | 0.26682 | standard |
| 7.63948 | 0.243846 | standard |
| -0.984184 | 6.125e-06 | standard |
| 3.65233 | 1.0 | standard |
| 6.94006 | 0.035859 | standard |
| 7.09355 | 0.215931 | standard |
| 7.22754 | 0.783088 | standard |
| 7.41563 | 0.389305 | standard |
| 7.55391 | 0.301629 | standard |
| 7.70722 | 0.11615 | standard |
| 7.74287 | 0.112694 | standard |
| -0.988747 | 3.125e-06 | standard |
| 3.65233 | 1.0 | standard |
| 7.00741 | 0.039871 | standard |
| 7.13028 | 0.233659 | standard |
| 7.22578 | 0.76453 | standard |
| 7.36423 | 0.14419 | standard |
| 7.4405 | 0.261132 | standard |
| 7.5024 | 0.167785 | standard |
| 7.57452 | 0.309589 | standard |
| 7.63074 | 0.144416 | standard |
| 7.69879 | 0.136668 | standard |
| -0.781249 | 1.125e-06 | standard |
| 3.65233 | 1.0 | standard |
| 7.04426 | 0.0453113 | standard |
| 7.13331 | 0.209148 | standard |
| 7.19434 | 0.411307 | standard |
| 7.22284 | 0.750829 | standard |
| 7.33596 | 0.102573 | standard |
| 7.38878 | 0.053702 | standard |
| 7.45181 | 0.243225 | standard |
| 7.54124 | 0.175713 | standard |
| 7.59082 | 0.242241 | standard |
| 7.67925 | 0.154826 | standard |
| -0.821122 | 1.125e-06 | standard |
| 3.65233 | 1.0 | standard |
| 7.05692 | 0.048828 | standard |
| 7.13653 | 0.206328 | standard |
| 7.1944 | 0.404165 | standard |
| 7.22207 | 0.70827 | standard |
| 7.3261 | 0.09515 | standard |
| 7.39726 | 0.0427684 | standard |
| 7.45569 | 0.242287 | standard |
| 7.53463 | 0.182344 | standard |
| 7.59508 | 0.241862 | standard |
| 7.67312 | 0.161861 | standard |
| -0.860995 | 1.125e-06 | standard |
| 3.65233 | 1.0 | standard |
| 7.06717 | 0.0527295 | standard |
| 7.13929 | 0.206725 | standard |
| 7.19472 | 0.400851 | standard |
| 7.22234 | 0.659845 | standard |
| 7.31764 | 0.0935432 | standard |
| 7.40281 | 0.0353311 | standard |
| 7.45884 | 0.242954 | standard |
| 7.52993 | 0.178819 | standard |
| 7.59818 | 0.240111 | standard |
| 7.66838 | 0.169346 | standard |
| 3.65233 | 1.0 | standard |
| 7.11355 | 0.0790393 | standard |
| 7.15034 | 0.225819 | standard |
| 7.18639 | 0.232298 | standard |
| 7.20586 | 0.247794 | standard |
| 7.22305 | 0.549568 | standard |
| 7.24041 | 0.290276 | standard |
| 7.27774 | 0.104009 | standard |
| 7.41749 | 0.00738612 | standard |
| 7.47327 | 0.272821 | standard |
| 7.50888 | 0.222611 | standard |
| 7.55526 | 0.0108123 | standard |
| 7.61312 | 0.267225 | standard |
| 7.64874 | 0.224086 | standard |
| 3.65233 | 1.0 | standard |
| 7.12823 | 0.0932648 | standard |
| 7.15295 | 0.242268 | standard |
| 7.17749 | 0.189225 | standard |
| 7.22296 | 0.547744 | standard |
| 7.23933 | 0.276669 | standard |
| 7.26418 | 0.112702 | standard |
| 7.47828 | 0.27402 | standard |
| 7.5027 | 0.238804 | standard |
| 7.61823 | 0.268705 | standard |
| 7.64263 | 0.238758 | standard |
| 3.65233 | 1.0 | standard |
| 7.13529 | 0.101611 | standard |
| 7.15391 | 0.248417 | standard |
| 7.17249 | 0.172914 | standard |
| 7.22279 | 0.594098 | standard |
| 7.23882 | 0.269391 | standard |
| 7.25742 | 0.116884 | standard |
| 7.48106 | 0.270365 | standard |
| 7.49953 | 0.243491 | standard |
| 7.62101 | 0.266828 | standard |
| 7.63948 | 0.243845 | standard |
| 3.65233 | 1.0 | standard |
| 7.1395 | 0.107009 | standard |
| 7.15439 | 0.251171 | standard |
| 7.16931 | 0.163789 | standard |
| 7.22317 | 0.644898 | standard |
| 7.23853 | 0.264889 | standard |
| 7.25351 | 0.119388 | standard |
| 7.48277 | 0.267668 | standard |
| 7.49763 | 0.246023 | standard |
| 7.62274 | 0.264897 | standard |
| 7.63758 | 0.246595 | standard |
| 3.65233 | 1.0 | standard |
| 7.14217 | 0.110662 | standard |
| 7.15465 | 0.252826 | standard |
| 7.16708 | 0.157945 | standard |
| 7.22313 | 0.550417 | standard |
| 7.23839 | 0.262471 | standard |
| 7.25084 | 0.121187 | standard |
| 7.48396 | 0.265623 | standard |
| 7.49634 | 0.247541 | standard |
| 7.62391 | 0.263574 | standard |
| 7.63632 | 0.248159 | standard |
| 3.65233 | 1.0 | standard |
| 7.1441 | 0.113556 | standard |
| 7.1548 | 0.253955 | standard |
| 7.16545 | 0.153856 | standard |
| 7.22306 | 0.521067 | standard |
| 7.22731 | 0.211504 | standard |
| 7.23831 | 0.260932 | standard |
| 7.24896 | 0.120885 | standard |
| 7.4848 | 0.264359 | standard |
| 7.49545 | 0.24861 | standard |
| 7.62477 | 0.259313 | standard |
| 7.63545 | 0.25283 | standard |
| 3.65233 | 1.0 | standard |
| 7.1449 | 0.114686 | standard |
| 7.15488 | 0.254075 | standard |
| 7.16481 | 0.152395 | standard |
| 7.22305 | 0.515082 | standard |
| 7.22816 | 0.189022 | standard |
| 7.23828 | 0.26015 | standard |
| 7.24822 | 0.121624 | standard |
| 7.48514 | 0.26352 | standard |
| 7.4951 | 0.249261 | standard |
| 7.62515 | 0.259324 | standard |
| 7.63505 | 0.25312 | standard |
| 3.65233 | 1.0 | standard |
| 7.14559 | 0.117326 | standard |
| 7.1549 | 0.252897 | standard |
| 7.16426 | 0.150766 | standard |
| 7.22305 | 0.511482 | standard |
| 7.22882 | 0.176058 | standard |
| 7.23823 | 0.259632 | standard |
| 7.24757 | 0.122023 | standard |
| 7.48544 | 0.26135 | standard |
| 7.49475 | 0.25101 | standard |
| 7.62544 | 0.2591 | standard |
| 7.63476 | 0.253305 | standard |
| 3.65233 | 1.0 | standard |
| 7.14668 | 0.118852 | standard |
| 7.15498 | 0.254788 | standard |
| 7.16327 | 0.146885 | standard |
| 7.22304 | 0.50742 | standard |
| 7.22986 | 0.161852 | standard |
| 7.23821 | 0.259016 | standard |
| 7.24653 | 0.123039 | standard |
| 7.48594 | 0.259052 | standard |
| 7.49421 | 0.253361 | standard |
| 7.62594 | 0.258736 | standard |
| 7.63426 | 0.253559 | standard |
| 3.65233 | 1.0 | standard |
| 7.14712 | 0.119314 | standard |
| 7.155 | 0.255141 | standard |
| 7.16287 | 0.145973 | standard |
| 7.22304 | 0.506238 | standard |
| 7.23027 | 0.158951 | standard |
| 7.23819 | 0.257155 | standard |
| 7.24604 | 0.123477 | standard |
| 7.48614 | 0.25876 | standard |
| 7.49401 | 0.253439 | standard |
| 7.62614 | 0.25739 | standard |
| 7.63401 | 0.254572 | standard |
| 3.65233 | 1.0 | standard |
| 7.14757 | 0.12016 | standard |
| 7.155 | 0.255261 | standard |
| 7.16253 | 0.14539 | standard |
| 7.22304 | 0.505279 | standard |
| 7.23067 | 0.155664 | standard |
| 7.23816 | 0.257968 | standard |
| 7.24564 | 0.122687 | standard |
| 7.48633 | 0.258563 | standard |
| 7.49381 | 0.253558 | standard |
| 7.62634 | 0.256286 | standard |
| 7.63376 | 0.256281 | standard |
| 3.65233 | 1.0 | standard |
| 7.14826 | 0.121075 | standard |
| 7.15507 | 0.255074 | standard |
| 7.16183 | 0.144233 | standard |
| 7.22304 | 0.503961 | standard |
| 7.23137 | 0.151081 | standard |
| 7.23811 | 0.258065 | standard |
| 7.24495 | 0.123376 | standard |
| 7.48668 | 0.258763 | standard |
| 7.49347 | 0.253274 | standard |
| 7.62663 | 0.256132 | standard |
| 7.63347 | 0.256128 | standard |
| 3.65233 | 1.0 | standard |
| 7.14935 | 0.123717 | standard |
| 7.1551 | 0.25572 | standard |
| 7.16084 | 0.141073 | standard |
| 7.22304 | 0.502512 | standard |
| 7.23237 | 0.14572 | standard |
| 7.23811 | 0.257464 | standard |
| 7.24386 | 0.124606 | standard |
| 7.48718 | 0.256194 | standard |
| 7.49292 | 0.256194 | standard |
| 7.62718 | 0.255332 | standard |
| 7.63297 | 0.256082 | standard |