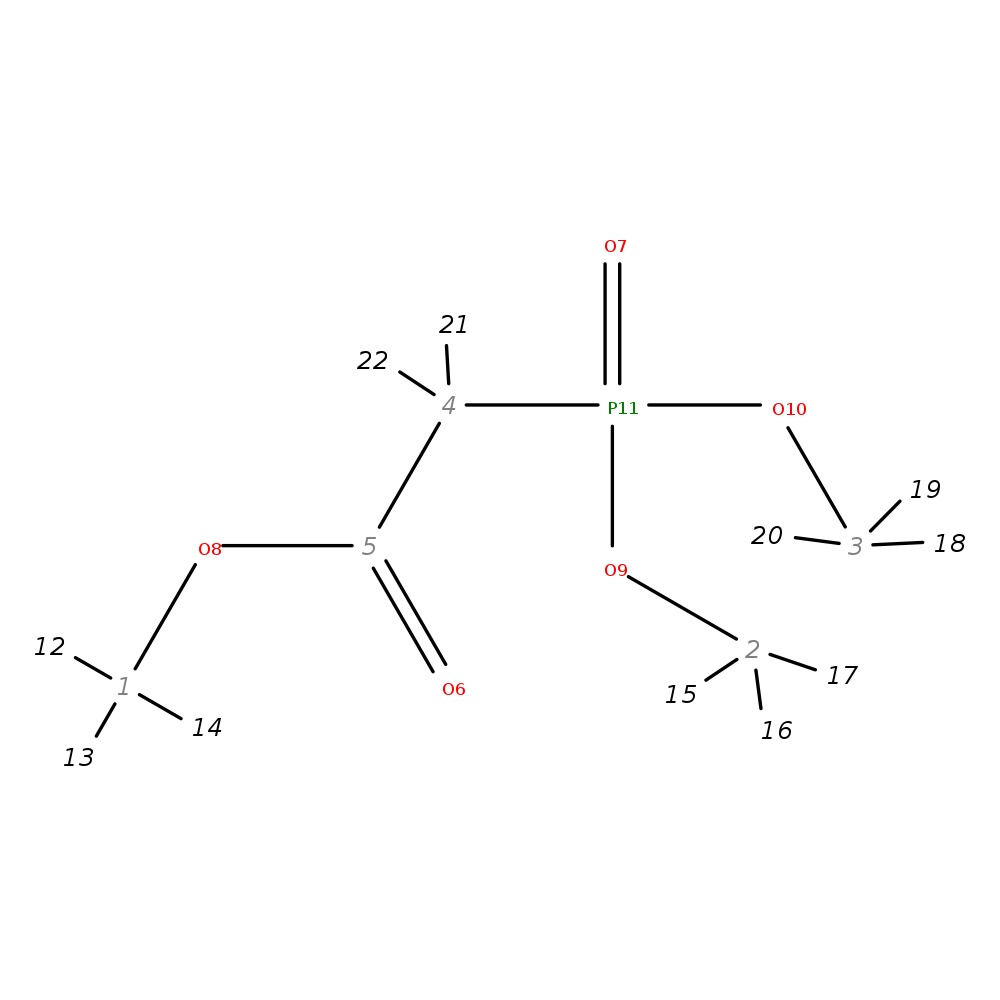
Trimethyl phosphonoacetate
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.00812 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C5H11O5P/c1-8-5(6)4-11(7,9-2)10-3/h4H2,1-3H3 | |
| Note 1 | Oscar Li |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Trimethylacetyl phosphate | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 3.808 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 3.808 | -12.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 3.808 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 3.837 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 3.837 | -12.4 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 3.837 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 3.837 | -12.4 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.837 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.837 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.769 | -12.4 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.769 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.76868 | 0.339611 | standard |
| 3.80835 | 0.505923 | standard |
| 3.83662 | 1.0 | standard |
| 3.83323 | 1.0 | standard |
| 3.77064 | 0.407887 | standard |
| 3.83563 | 1.0 | standard |
| 3.7693 | 0.378559 | standard |
| 3.80975 | 0.631902 | standard |
| 3.83625 | 1.0 | standard |
| 3.76907 | 0.370419 | standard |
| 3.80923 | 0.607786 | standard |
| 3.83637 | 1.00002 | standard |
| 3.76894 | 0.364488 | standard |
| 3.80893 | 0.589624 | standard |
| 3.83645 | 1.00001 | standard |
| 3.76869 | 0.34477 | standard |
| 3.80839 | 0.524477 | standard |
| 3.83661 | 1.00001 | standard |
| 3.76868 | 0.340959 | standard |
| 3.80836 | 0.510844 | standard |
| 3.83662 | 1.0 | standard |
| 3.76868 | 0.339612 | standard |
| 3.80835 | 0.505927 | standard |
| 3.83662 | 1.0 | standard |
| 3.76868 | 0.338986 | standard |
| 3.80835 | 0.503623 | standard |
| 3.83662 | 1.0 | standard |
| 3.76868 | 0.338646 | standard |
| 3.80835 | 0.502364 | standard |
| 3.83662 | 1.0 | standard |
| 3.76868 | 0.33844 | standard |
| 3.80835 | 0.501603 | standard |
| 3.83662 | 1.0 | standard |
| 3.76868 | 0.338367 | standard |
| 3.80835 | 0.501331 | standard |
| 3.83662 | 1.0 | standard |
| 3.76868 | 0.338307 | standard |
| 3.80835 | 0.501108 | standard |
| 3.83662 | 1.0 | standard |
| 3.76868 | 0.338215 | standard |
| 3.80835 | 0.500768 | standard |
| 3.83662 | 1.0 | standard |
| 3.76868 | 0.33818 | standard |
| 3.80835 | 0.500636 | standard |
| 3.83662 | 1.0 | standard |
| 3.76868 | 0.33815 | standard |
| 3.80835 | 0.500524 | standard |
| 3.83662 | 1.0 | standard |
| 3.76868 | 0.338101 | standard |
| 3.80835 | 0.500344 | standard |
| 3.83662 | 1.0 | standard |
| 3.76868 | 0.338036 | standard |
| 3.80835 | 0.5001 | standard |
| 3.83662 | 1.0 | standard |