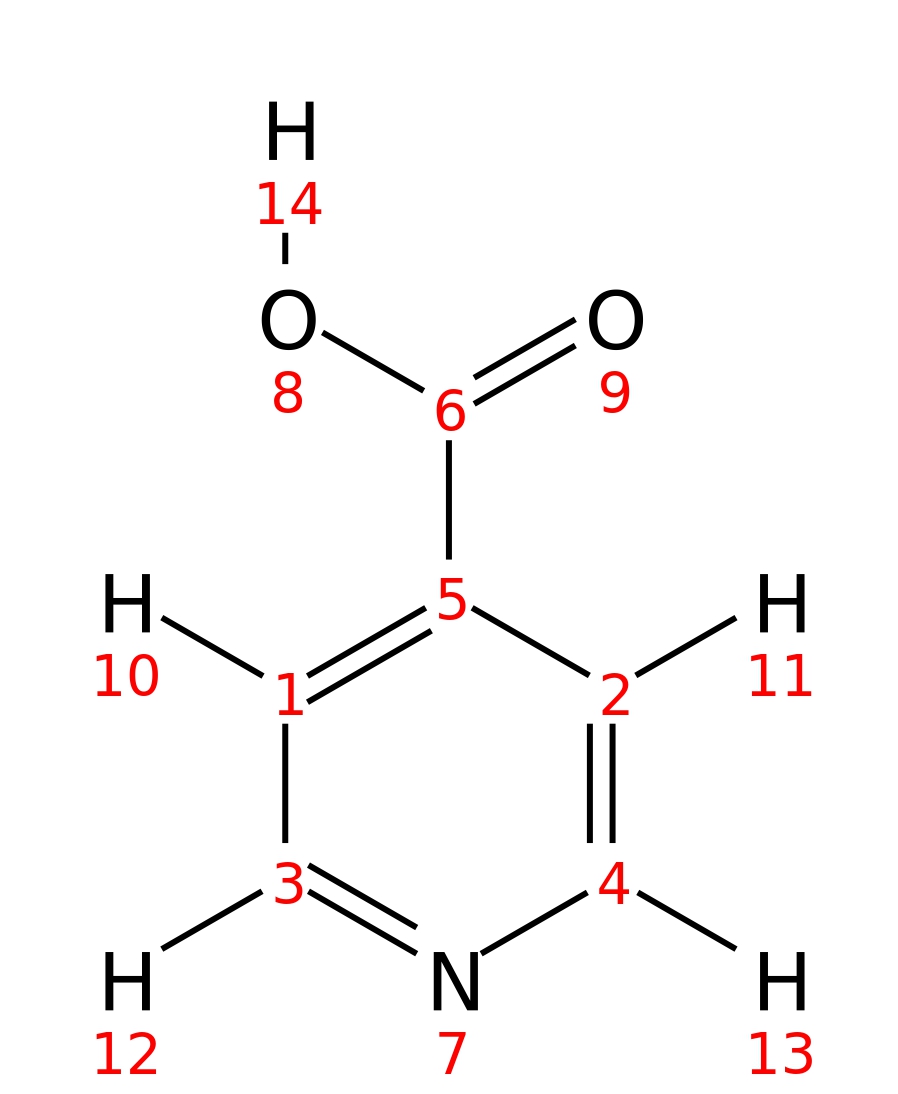
Isonicotinic-acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.01586 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H5NO2/c8-6(9)5-1-3-7-4-2-5/h1-4H,(H,8,9) | |
| Note 1 | 10,12?11,13 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Isonicotinic acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 12 | 10 | 11 | 13 | |
|---|---|---|---|---|
| 12 | 7.669 | 4.541 | 2.12 | 0 |
| 10 | 0 | 8.659 | 0 | 2.037 |
| 11 | 0 | 0 | 7.669 | 6.974 |
| 13 | 0 | 0 | 0 | 8.659 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| -0.271629 | 1.125e-06 | standard |
| 7.66148 | 0.992705 | standard |
| 7.67568 | 1.00028 | standard |
| 8.65183 | 0.995806 | standard |
| 8.66599 | 0.991602 | standard |
| -0.992516 | 9.8125e-05 | standard |
| 7.59223 | 0.741804 | standard |
| 7.73435 | 1.0 | standard |
| 8.59306 | 0.996068 | standard |
| 8.73498 | 0.737178 | standard |
| -0.979225 | 4.5125e-05 | standard |
| 7.61892 | 0.818834 | standard |
| 7.71354 | 1.0 | standard |
| 8.61395 | 0.99558 | standard |
| 8.70843 | 0.815043 | standard |
| -0.975257 | 2.6125e-05 | standard |
| 7.63177 | 0.861052 | standard |
| 7.70277 | 1.00001 | standard |
| 8.62475 | 0.995517 | standard |
| 8.69557 | 0.856576 | standard |
| -0.98468 | 2.1125e-05 | standard |
| 7.63604 | 0.875171 | standard |
| 7.69911 | 1.0 | standard |
| 8.6284 | 0.994984 | standard |
| 8.69138 | 0.870898 | standard |
| -0.972778 | 1.7125e-05 | standard |
| 7.6394 | 0.887047 | standard |
| 7.69615 | 1.0 | standard |
| 8.63132 | 0.995711 | standard |
| 8.688 | 0.882452 | standard |
| -0.779761 | 4.125e-06 | standard |
| 7.65423 | 0.957341 | standard |
| 7.68263 | 1.00005 | standard |
| 8.64489 | 0.996477 | standard |
| 8.67318 | 0.957234 | standard |
| -0.738797 | 2.125e-06 | standard |
| 7.65906 | 0.974899 | standard |
| 7.67801 | 1.0 | standard |
| 8.64948 | 0.99725 | standard |
| 8.66838 | 0.973614 | standard |
| -0.270836 | 1.125e-06 | standard |
| 7.66148 | 0.993104 | standard |
| 7.67568 | 1.00028 | standard |
| 8.65183 | 0.996214 | standard |
| 8.66599 | 0.992007 | standard |
| -0.663812 | 1.125e-06 | standard |
| 7.66291 | 0.995431 | standard |
| 7.67427 | 1.00019 | standard |
| 8.65319 | 0.989485 | standard |
| 8.66456 | 0.983447 | standard |
| 7.6639 | 0.998787 | standard |
| 7.67333 | 1.00009 | standard |
| 8.65412 | 0.993023 | standard |
| 8.6636 | 0.992693 | standard |
| 7.66454 | 0.997021 | standard |
| 7.67266 | 1.00001 | standard |
| 8.65481 | 0.99625 | standard |
| 8.66291 | 0.996256 | standard |
| 7.66482 | 0.994249 | standard |
| 7.6724 | 1.00074 | standard |
| 8.65507 | 0.996068 | standard |
| 8.66261 | 0.991304 | standard |
| 7.66507 | 0.99624 | standard |
| 7.67219 | 1.00043 | standard |
| 8.65533 | 0.9961 | standard |
| 8.66239 | 0.990061 | standard |
| 7.66548 | 0.990202 | standard |
| 7.67176 | 1.00001 | standard |
| 8.65569 | 0.989967 | standard |
| 8.66198 | 0.9895 | standard |
| 7.66564 | 0.994788 | standard |
| 7.67162 | 1.00013 | standard |
| 8.65587 | 1.00022 | standard |
| 8.66184 | 0.996498 | standard |
| 7.6658 | 0.999104 | standard |
| 7.67147 | 1.00003 | standard |
| 8.65603 | 0.980758 | standard |
| 8.66169 | 0.980905 | standard |
| 7.66608 | 1.00045 | standard |
| 7.67119 | 0.992828 | standard |
| 8.65627 | 0.988223 | standard |
| 8.66143 | 0.984213 | standard |
| 7.66643 | 1.00037 | standard |
| 7.67083 | 1.0012 | standard |
| 8.65666 | 0.978537 | standard |
| 8.66102 | 0.991973 | standard |