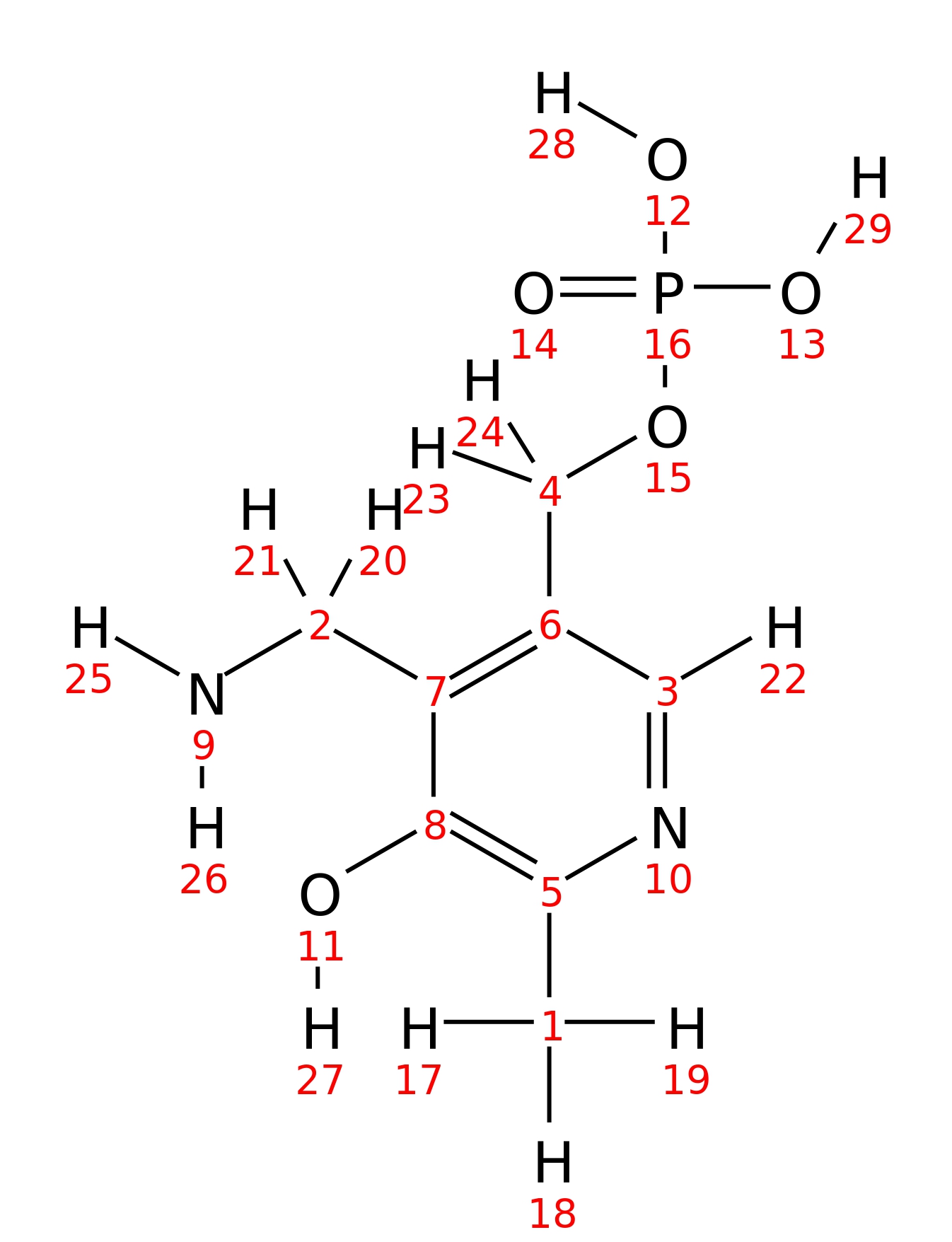
Pyridoxamine-5-phosphate
Simulation outputs:
|
Parameter | Value | |||||
| Field strength | 400.13(MHz) | ||||||
| RMSD of the fit | 0.03191 | ||||||
| Temperature | 298 K | ||||||
| pH | 7.4 | ||||||
| InChI | InChI=1S/C8H13N2O5P/c1-5-8(11)7(2-9)6(3-10-5)4-15-16(12,13)14/h3,11H,2,4,9H2,1H3,(H2,12,13,14) | ||||||
| Note 1 | overlap with water 23,24 inaccurate shift | ||||||
| Note 2 | estimated 31P Jcoupling to 23,24 from 2D HSQC (7Hz) | ||||||
| Note 3 | could change temperature of 1D | ||||||
| Note 4 | |||||||
| Additional coupling constants |
|
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Pyridoxamine phosphate | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 17 | 18 | 19 | 22 | 20 | 21 | 23 | 24 | |
|---|---|---|---|---|---|---|---|---|
| 17 | 2.461 | -14.9 | -14.9 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 2.461 | -14.9 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 2.461 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 7.645 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 4.315 | -12.4 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 4.315 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 4.84 | -12.4 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.84 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.888688 | standard |
| 4.83131 | 0.472642 | standard |
| 4.84866 | 0.472638 | standard |
| 7.64537 | 0.444435 | standard |
| 2.4612 | 1.0 | standard |
| 4.31472 | 0.895529 | standard |
| 4.75308 | 0.481751 | standard |
| 4.92675 | 0.477151 | standard |
| 7.64537 | 0.444341 | standard |
| 2.4612 | 1.0 | standard |
| 4.31471 | 0.891313 | standard |
| 4.7821 | 0.476118 | standard |
| 4.89784 | 0.474784 | standard |
| 7.64537 | 0.444366 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.890416 | standard |
| 4.79659 | 0.474471 | standard |
| 4.88338 | 0.473912 | standard |
| 7.64537 | 0.44442 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.889845 | standard |
| 4.80141 | 0.473793 | standard |
| 4.87856 | 0.473401 | standard |
| 7.64537 | 0.444351 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.890043 | standard |
| 4.80527 | 0.473813 | standard |
| 4.8747 | 0.473528 | standard |
| 7.64537 | 0.444499 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.88926 | standard |
| 4.82263 | 0.472756 | standard |
| 4.85735 | 0.47272 | standard |
| 7.64537 | 0.444438 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.888767 | standard |
| 4.82842 | 0.472636 | standard |
| 4.85156 | 0.472625 | standard |
| 7.64537 | 0.444386 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.88896 | standard |
| 4.83131 | 0.472603 | standard |
| 4.84867 | 0.4726 | standard |
| 7.64537 | 0.444415 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.888772 | standard |
| 4.83304 | 0.472646 | standard |
| 4.84693 | 0.472644 | standard |
| 7.64537 | 0.444315 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.888872 | standard |
| 4.8342 | 0.47251 | standard |
| 4.84577 | 0.472509 | standard |
| 7.64537 | 0.444423 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.888736 | standard |
| 4.83503 | 0.472457 | standard |
| 4.84495 | 0.472457 | standard |
| 7.64537 | 0.444397 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.888825 | standard |
| 4.83536 | 0.472733 | standard |
| 4.84462 | 0.472732 | standard |
| 7.64537 | 0.444515 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.888536 | standard |
| 4.83565 | 0.472438 | standard |
| 4.84433 | 0.472438 | standard |
| 7.64537 | 0.444257 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.888704 | standard |
| 4.83613 | 0.472477 | standard |
| 4.84385 | 0.472477 | standard |
| 7.64537 | 0.444302 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.888846 | standard |
| 4.83633 | 0.472866 | standard |
| 4.84364 | 0.472866 | standard |
| 7.64537 | 0.444538 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.88866 | standard |
| 4.83652 | 0.472451 | standard |
| 4.84346 | 0.472451 | standard |
| 7.64537 | 0.444377 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.888716 | standard |
| 4.83683 | 0.472517 | standard |
| 4.84314 | 0.472517 | standard |
| 7.64537 | 0.444341 | standard |
| 2.4612 | 1.0 | standard |
| 4.3147 | 0.888756 | standard |
| 4.83732 | 0.472586 | standard |
| 4.84266 | 0.472586 | standard |
| 7.64537 | 0.44431 | standard |