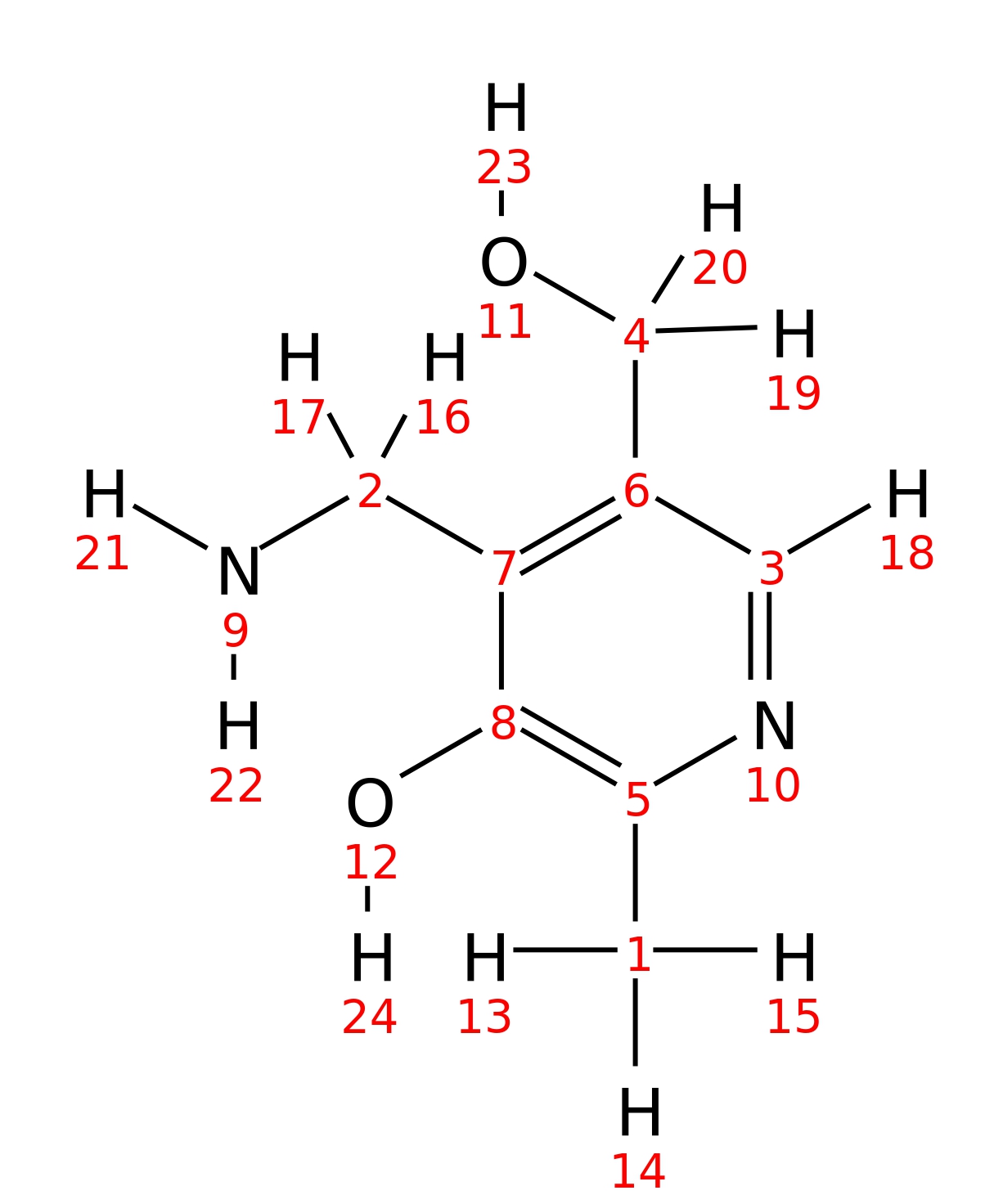
Pyridoxamine
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.00805 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H12N2O2/c1-5-8(12)7(2-9)6(4-11)3-10-5/h3,11-12H,2,4,9H2,1H3 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Pyridoxamine | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 13 | 14 | 15 | 18 | 19 | 20 | 16 | 17 | |
|---|---|---|---|---|---|---|---|---|
| 13 | 2.46 | -14.9 | -14.9 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 2.46 | -14.9 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 2.46 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 7.609 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 4.299 | -12.4 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 4.299 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 4.702 | -12.4 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.702 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.666675 | standard |
| 4.70192 | 0.666995 | standard |
| 7.60868 | 0.332874 | standard |
| 2.45962 | 1.0 | standard |
| 4.29945 | 0.672595 | standard |
| 4.70188 | 0.672493 | standard |
| 7.60868 | 0.333013 | standard |
| 2.45962 | 1.0 | standard |
| 4.29943 | 0.668946 | standard |
| 4.70191 | 0.669054 | standard |
| 7.60868 | 0.332955 | standard |
| 2.45962 | 1.0 | standard |
| 4.29943 | 0.667779 | standard |
| 4.70192 | 0.667628 | standard |
| 7.60868 | 0.333044 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.667617 | standard |
| 4.70192 | 0.667522 | standard |
| 7.60868 | 0.332954 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.667681 | standard |
| 4.70192 | 0.667725 | standard |
| 7.60868 | 0.332911 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.666785 | standard |
| 4.70192 | 0.666806 | standard |
| 7.60868 | 0.333211 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.666697 | standard |
| 4.70192 | 0.666722 | standard |
| 7.60868 | 0.333267 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.666678 | standard |
| 4.70192 | 0.666364 | standard |
| 7.60868 | 0.333283 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.666674 | standard |
| 4.70192 | 0.666454 | standard |
| 7.60868 | 0.332982 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.666274 | standard |
| 4.70192 | 0.666375 | standard |
| 7.60868 | 0.332779 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.666614 | standard |
| 4.70192 | 0.666728 | standard |
| 7.60868 | 0.333407 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.666755 | standard |
| 4.70192 | 0.666646 | standard |
| 7.60868 | 0.333251 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.666093 | standard |
| 4.70192 | 0.666371 | standard |
| 7.60868 | 0.332977 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.666331 | standard |
| 4.70192 | 0.666162 | standard |
| 7.60868 | 0.333036 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.667152 | standard |
| 4.70192 | 0.666696 | standard |
| 7.60868 | 0.333304 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.666879 | standard |
| 4.70192 | 0.666895 | standard |
| 7.60868 | 0.333198 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.6666 | standard |
| 4.70192 | 0.666303 | standard |
| 7.60868 | 0.33287 | standard |
| 2.45962 | 1.0 | standard |
| 4.29942 | 0.665943 | standard |
| 4.70192 | 0.666038 | standard |
| 7.60868 | 0.333122 | standard |