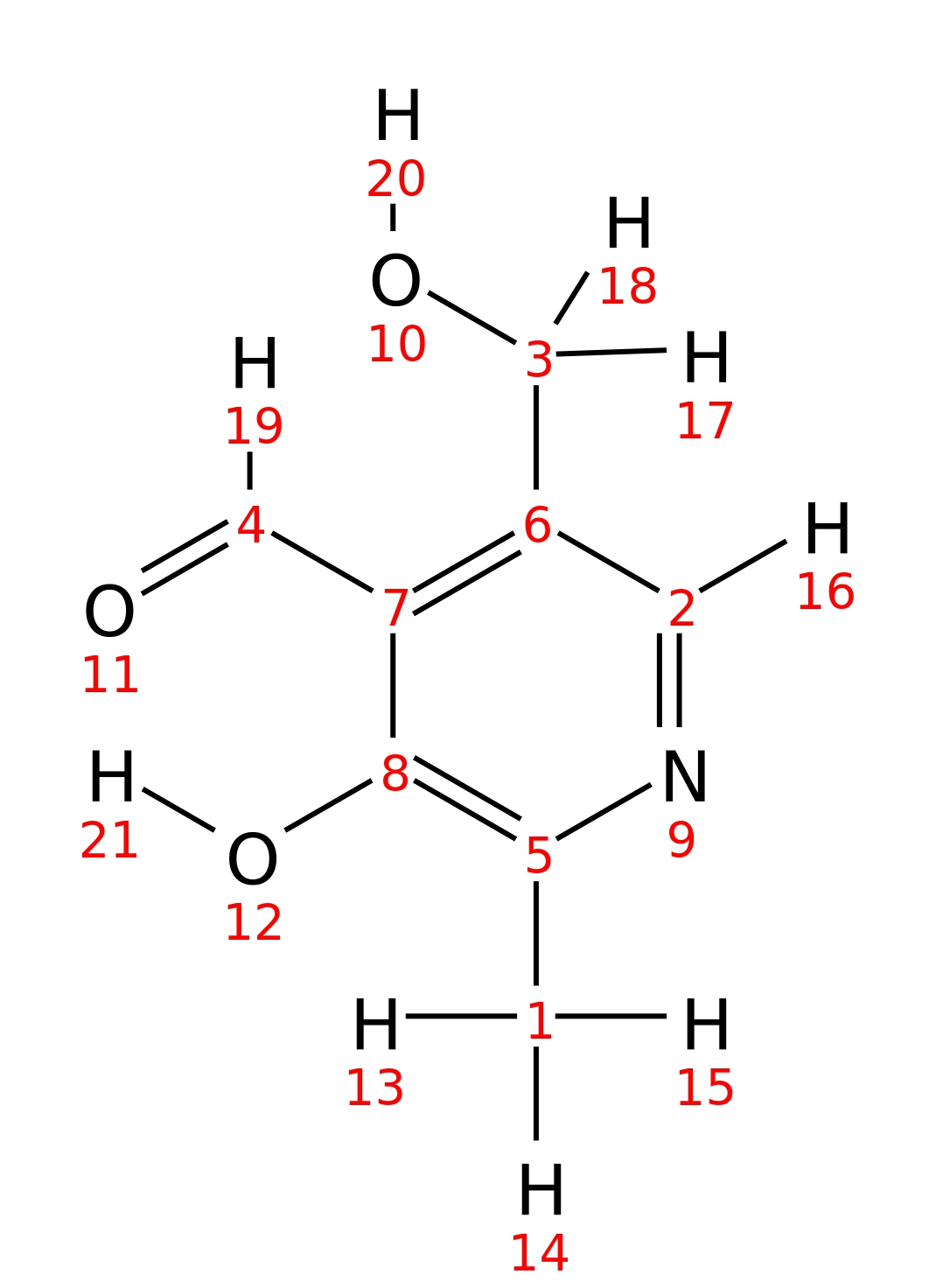
Pyridoxal
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.01694 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H9NO3/c1-5-8(12)7(4-11)6(3-10)2-9-5/h2,4,10,12H,3H2,1H3 | |
| Note 1 | qm - hydrated aldehyde |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Pyridoxal | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 13 | 14 | 15 | 19 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|
| 13 | 2.469 | -14.9 | -14.9 | 0 | 0 | 0 | 0 |
| 14 | 0 | 2.469 | -14.9 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 2.469 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 6.553 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 7.578 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 5.241 | -13.009 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 5.055 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.46884 | 1.0 | standard |
| 5.03699 | 0.141936 | standard |
| 5.0695 | 0.198092 | standard |
| 5.22652 | 0.198092 | standard |
| 5.25903 | 0.141937 | standard |
| 6.55284 | 0.333341 | standard |
| 7.57823 | 0.333336 | standard |
| 2.46884 | 1.0 | standard |
| 4.80086 | 0.0330091 | standard |
| 5.14821 | 0.478126 | standard |
| 5.49556 | 0.0333091 | standard |
| 6.55284 | 0.334274 | standard |
| 7.57823 | 0.333777 | standard |
| 2.46884 | 1.0 | standard |
| 4.89737 | 0.0494661 | standard |
| 5.11521 | 0.341752 | standard |
| 5.18081 | 0.34169 | standard |
| 5.39865 | 0.0495671 | standard |
| 6.55284 | 0.33382 | standard |
| 7.57823 | 0.333596 | standard |
| 2.46884 | 1.0 | standard |
| 4.94329 | 0.0649871 | standard |
| 5.10587 | 0.295701 | standard |
| 5.19015 | 0.295708 | standard |
| 5.3528 | 0.06502 | standard |
| 6.55284 | 0.33344 | standard |
| 7.57823 | 0.333315 | standard |
| 2.46884 | 1.0 | standard |
| 4.95787 | 0.0719193 | standard |
| 5.10242 | 0.282735 | standard |
| 5.1937 | 0.282743 | standard |
| 5.33815 | 0.0719575 | standard |
| 6.55284 | 0.333572 | standard |
| 7.57823 | 0.333473 | standard |
| 2.46884 | 1.0 | standard |
| 4.96933 | 0.07824 | standard |
| 5.09936 | 0.27247 | standard |
| 5.19666 | 0.272476 | standard |
| 5.3267 | 0.078262 | standard |
| 6.55284 | 0.333524 | standard |
| 7.57823 | 0.333444 | standard |
| 2.46884 | 1.0 | standard |
| 5.01669 | 0.11664 | standard |
| 5.0817 | 0.224861 | standard |
| 5.21442 | 0.224862 | standard |
| 5.27943 | 0.116643 | standard |
| 6.55284 | 0.333381 | standard |
| 7.57823 | 0.333361 | standard |
| 2.46884 | 1.0 | standard |
| 5.03045 | 0.133109 | standard |
| 5.07386 | 0.207254 | standard |
| 5.22226 | 0.207266 | standard |
| 5.26557 | 0.133128 | standard |
| 6.55284 | 0.333355 | standard |
| 7.57823 | 0.333346 | standard |
| 2.46884 | 1.0 | standard |
| 5.03699 | 0.141924 | standard |
| 5.0695 | 0.197787 | standard |
| 5.22653 | 0.197788 | standard |
| 5.25903 | 0.141925 | standard |
| 6.55284 | 0.333073 | standard |
| 7.57823 | 0.333068 | standard |
| 2.46884 | 1.0 | standard |
| 5.04075 | 0.147309 | standard |
| 5.06672 | 0.192528 | standard |
| 5.2293 | 0.192529 | standard |
| 5.25527 | 0.14731 | standard |
| 6.55284 | 0.332898 | standard |
| 7.57823 | 0.332895 | standard |
| 2.46884 | 1.0 | standard |
| 5.04323 | 0.151055 | standard |
| 5.06484 | 0.188784 | standard |
| 5.23118 | 0.188759 | standard |
| 5.25289 | 0.151024 | standard |
| 6.55284 | 0.333338 | standard |
| 7.57823 | 0.333336 | standard |
| 2.46884 | 1.0 | standard |
| 5.04492 | 0.153514 | standard |
| 5.06345 | 0.186084 | standard |
| 5.23257 | 0.186054 | standard |
| 5.25121 | 0.153478 | standard |
| 6.55284 | 0.333138 | standard |
| 7.57823 | 0.333136 | standard |
| 2.46884 | 1.0 | standard |
| 5.04561 | 0.154738 | standard |
| 5.06295 | 0.184992 | standard |
| 5.23317 | 0.184993 | standard |
| 5.25051 | 0.154738 | standard |
| 6.55284 | 0.333337 | standard |
| 7.57823 | 0.333335 | standard |
| 2.46884 | 1.0 | standard |
| 5.0462 | 0.155607 | standard |
| 5.06246 | 0.183992 | standard |
| 5.23366 | 0.183992 | standard |
| 5.24992 | 0.155608 | standard |
| 6.55284 | 0.333216 | standard |
| 7.57823 | 0.333214 | standard |
| 2.46884 | 1.0 | standard |
| 5.04709 | 0.157219 | standard |
| 5.06157 | 0.182462 | standard |
| 5.23446 | 0.182462 | standard |
| 5.24893 | 0.157219 | standard |
| 6.55284 | 0.333336 | standard |
| 7.57823 | 0.333335 | standard |
| 2.46884 | 1.0 | standard |
| 5.04759 | 0.157891 | standard |
| 5.06127 | 0.18181 | standard |
| 5.23485 | 0.18181 | standard |
| 5.24853 | 0.157891 | standard |
| 6.55284 | 0.333335 | standard |
| 7.57823 | 0.333335 | standard |
| 2.46884 | 1.0 | standard |
| 5.04789 | 0.158488 | standard |
| 5.06087 | 0.181216 | standard |
| 5.23515 | 0.181216 | standard |
| 5.24813 | 0.158488 | standard |
| 6.55284 | 0.333335 | standard |
| 7.57823 | 0.333334 | standard |
| 2.46884 | 1.0 | standard |
| 5.04858 | 0.159516 | standard |
| 5.06038 | 0.180186 | standard |
| 5.23565 | 0.180138 | standard |
| 5.24754 | 0.159462 | standard |
| 6.55284 | 0.333335 | standard |
| 7.57823 | 0.333334 | standard |
| 2.46884 | 1.0 | standard |
| 5.04947 | 0.161071 | standard |
| 5.05948 | 0.178542 | standard |
| 5.23654 | 0.178542 | standard |
| 5.24655 | 0.161071 | standard |
| 6.55284 | 0.333331 | standard |
| 7.57823 | 0.33333 | standard |